Identification of Haemophilus Species and the HACEK Group of Organisms
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Reporting of Diseases and Conditions Regulation, Amendment, M.R. 289/2014
THE PUBLIC HEALTH ACT LOI SUR LA SANTÉ PUBLIQUE (C.C.S.M. c. P210) (c. P210 de la C.P.L.M.) Reporting of Diseases and Conditions Règlement modifiant le Règlement sur la Regulation, amendment déclaration de maladies et d'affections Regulation 289/2014 Règlement 289/2014 Registered December 23, 2014 Date d'enregistrement : le 23 décembre 2014 Manitoba Regulation 37/2009 amended Modification du R.M. 37/2009 1 The Reporting of Diseases and 1 Le présent règlement modifie le Conditions Regulation , Manitoba Règlement sur la déclaration de maladies et Regulation 37/2009, is amended by this d'affections , R.M. 37/2009. regulation. 2 Schedules A and B are replaced with 2 Les annexes A et B sont remplacées Schedules A and B to this regulation. par les annexes A et B du présent règlement. Coming into force Entrée en vigueur 3 This regulation comes into force on 3 Le présent règlement entre en vigueur January 1, 2015, or on the day it is registered le 1 er janvier 2015 ou à la date de son under The Statutes and Regulations Act , enregistrement en vertu de Loi sur les textes whichever is later. législatifs et réglementaires , si cette date est postérieure. December 19, 2014 Minister of Health/La ministre de la Santé, 19 décembre 2014 Sharon Blady 1 SCHEDULE A (Section 1) 1 The following diseases are diseases requiring contact notification in accordance with the disease-specific protocol. Common name Scientific or technical name of disease or its infectious agent Chancroid Haemophilus ducreyi Chlamydia Chlamydia trachomatis (including Lymphogranuloma venereum (LGV) serovars) Gonorrhea Neisseria gonorrhoeae HIV Human immunodeficiency virus Syphilis Treponema pallidum subspecies pallidum Tuberculosis Mycobacterium tuberculosis Mycobacterium africanum Mycobacterium canetti Mycobacterium caprae Mycobacterium microti Mycobacterium pinnipedii Mycobacterium bovis (excluding M. -

Burkholderia Cepacia Complex As Human Pathogens1
Journal of Nematology 35(2):212–217. 2003. © The Society of Nematologists 2003. Burkholderia cepacia Complex as Human Pathogens1 John J. LiPuma2 Abstract: Although sporadic human infection due to Burkholderia cepacia has been reported for many years, it has been only during the past few decades that species within the B. cepacia complex have emerged as significant opportunistic human pathogens. Individuals with cystic fibrosis, the most common inherited genetic disease in Caucasian populations, or chronic granulomatous disease, a primary immunodeficiency, are particularly at risk of life-threatening infection. Despite advances in our understanding of the taxonomy, microbiology, and epidemiology of B. cepacia complex, much remains unknown regarding specific human virulence factors. The broad-spectrum antimicrobial resistance demonstrated by most strains limits current therapy of infection. Recent research efforts are aimed at a better appreciation of the pathogenesis of human infection and the development of novel therapeutic and prophylactic strategies. Key words: Burkholderia cepacia, cystic fibrosis, human infection. Until relatively recently, Burkholderia cepacia had been B. cepacia, possess other factors that remain to be elu- considered a phytopathogenic or saprophytic bacterial cidated that also mediate pathogenicity in this condi- species with little potential for human infection. How- tion (Speert et al., 1994). Fortunately, CGD is a rela- ever, reports of sporadic human infection have ap- tively rare disease, having an average annual incidence peared in the biomedical literature, generally describ- of approximately 1/200,000 live births in the United ing infection in persons with some underlying disease States; this means there are approximately 20 persons or debilitation (Dailey and Benner, 1968; Poe et al., with CGD born each year in the United States. -

BD-CS-057, REV 0 | AUGUST 2017 | Page 1
EXPLIFY RESPIRATORY PATHOGENS BY NEXT GENERATION SEQUENCING Limitations Negative results do not rule out viral, bacterial, or fungal infections. Targeted, PCR-based tests are generally more sensitive and are preferred when specific pathogens are suspected, especially for DNA viruses (Adenovirus, CMV, HHV6, HSV, and VZV), mycobacteria, and fungi. The analytical sensitivity of this test depends on the cellularity of the sample and the concentration of all microbes present. Analytical sensitivity is assessed using Internal Controls that are added to each sample. Sequencing data for Internal Controls is quantified. Samples with Internal Control values below the validated minimum may have reduced analytical sensitivity or contain inhibitors and are reported as ‘Reduced Analytical Sensitivity’. Additional respiratory pathogens to those reported cannot be excluded in samples with ‘Reduced Analytical Sensitivity’. Due to the complexity of next generation sequencing methodologies, there may be a risk of false-positive results. Contamination with organisms from the upper respiratory tract during specimen collection can also occur. The detection of viral, bacterial, and fungal nucleic acid does not imply organisms causing invasive infection. Results from this test need to be interpreted in conjunction with the clinical history, results of other laboratory tests, epidemiologic information, and other available data. Confirmation of positive results by an alternate method may be indicated in select cases. Validated Organisms BACTERIA Achromobacter -

Studies on Oral Colonization of Periodontopathogenic Bacterium
Studies on oral colonization of periodontopathogenic bacterium Eikenella corrodens 㸦ṑ࿘ཎᛶ⣽⳦ (LNHQHOODFRUURGHQV ࡢཱྀ⭍ෆᐃ╔㛵ࡍࡿ◊✲㸧 㻌 㻌 FARIHA JASIN MANSUR 2017 㻌 㻌 DEDICATED TO MY BELOVED PARENTS CONTENTS CONTENTS…………………………………………………………. 1 LIST OF ABBREVIATIONS ……………………………………. 2 CHAPTER 1: GENERAL INTRODUCTION …………………………………... 4 CHAPTER 2 .………………………………………………………. 11 2.1 ABSTRACT ……………………………………………………. 12 2.2 INTRODUCTION ……………………………………………... 13 2.3 MATERIALS AND METHODS ……………………………… 16 2.4 RESULTS AND DISCUSSION ……………………………….. 21 CHAPTER 3 ………………………………………………………... 31 3.1 ABSTRACT …………………………………………………….. 32 3.2 INTRODUCTION ……………………………………………… 33 3.3 MATERIALS AND METHODS ………………………………. 35 3.4 RESULTS AND DISCUSSION ………………………………... 38 CHAPTER 4: GENERAL CONCLUSION ………………………………………... 44 SUMMARY ………………………………………………………….. 50 JAPANESE SUMMARY ……………………………………………. 52 ACKNOWLEDGEMENTS …………………………………………. 54 REFERENCES ……………………………………………………….. 56 LIST OF PUBLICATIONS ………………………………………….. 65 㻝㻌 㻌 LIST OF ABBREVIATIONS CE Cell envelope GalNAc㻌㻌㻌㻌㻌㻌 N-acetyl-D-galactosamine g Gram g/L Gram/litre HA㻌㻌㻌㻌㻌㻌㻌㻌 Hemagglutination ∆hlyA hlyA-deficient strain H hour IL Interleukin IPTG Isopropyl β-D-1-thiogalactopyranoside LB㻌 㻌 㻌 㻌 Luria broth M Molar mM Milimolar min Minute mL Mililitre mg/mL Milligram/mililitre NaCl Sodium chloride ORF Open reading frame PBS Phosphate-buffered saline㻌 PCR Polymerase chain reaction pH㻌㻌 㻌 㻌㻌㻌㻌㻌㻌㻌Potential of hydrogen SDS–PAGE Sodium dodecyl sulfate polyacrylamide gel electrophoresis TSB Tryptic soy broth 㻞㻌 㻌 μL Microlitre μM Micromolar -

Reportable Disease Surveillance in Virginia, 2013
Reportable Disease Surveillance in Virginia, 2013 Marissa J. Levine, MD, MPH State Health Commissioner Report Production Team: Division of Surveillance and Investigation, Division of Disease Prevention, Division of Environmental Epidemiology, and Division of Immunization Virginia Department of Health Post Office Box 2448 Richmond, Virginia 23218 www.vdh.virginia.gov ACKNOWLEDGEMENT In addition to the employees of the work units listed below, the Office of Epidemiology would like to acknowledge the contributions of all those engaged in disease surveillance and control activities across the state throughout the year. We appreciate the commitment to public health of all epidemiology staff in local and district health departments and the Regional and Central Offices, as well as the conscientious work of nurses, environmental health specialists, infection preventionists, physicians, laboratory staff, and administrators. These persons report or manage disease surveillance data on an ongoing basis and diligently strive to control morbidity in Virginia. This report would not be possible without the efforts of all those who collect and follow up on morbidity reports. Divisions in the Virginia Department of Health Office of Epidemiology Disease Prevention Telephone: 804-864-7964 Environmental Epidemiology Telephone: 804-864-8182 Immunization Telephone: 804-864-8055 Surveillance and Investigation Telephone: 804-864-8141 TABLE OF CONTENTS INTRODUCTION Introduction ......................................................................................................................................1 -

( Eikenella Corrodens のクオラムセンシングと バイオフィルム形成の関連性に関する研究)
Studies on the relationship between quorum sensing and biofilm formation of Eikenella corrodens ( Eikenella corrodens のクオラムセンシングと バイオフィルム形成の関連性に関する研究) by MOHAMMAD MINNATUL KARIM Thesis Submitted to the United Graduate School of Agricultural Sciences Tottori University, Japan In partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY (PhD) The United Graduate School of Agricultural Sciences Tottori University, Japan September, 2013 1 DEDICATED TO MY BELOVED PARENTS 2 CONTENTS CONTENTS...............................................................................................ⅰ LIST OF ABBREVIATIONS...................................................................ⅱ GENERAL INTRODUCTION................................................................. 1 OBJECTIVES........................................................................................... 10 CHAPTER 1 ............................................................................................. 11 The Periodontopathogenic Bacterium Eikenella corrodens Produces an Autoinducer-2-Inactivating Enzyme 1.1 ABSTRACT ………………………………………………………… 12 1.2 INTRODUCTION …………………………………………………....13 1.3 MATERIALS AND METHODS ……………………………………..15 1.4 RESULTS …………………………………………………………… 19 1.5 DISCUSSION ………………………………………………………. 22 1.6 FIGURES …………………………………………………………….25 CHAPTER 2 …………………………………………………………… 32 LuxS affects biofilm maturation and detachment of the periodontopathogenic bacterium Eikenella corrodens 2.1 ABSTRACT …………………………………………………………. 33 2.2 INTRODUCTION …………………………………………………....34 -

Digestive Tract Infections •Skin & Soft Tissue Infections(SSTI)
EU Conference: Workshop #6 Macrolide Indications: •Ocular Infections •Stomatologic Infections •Digestive tract Infections •Skin & Soft Tissue Infections(SSTI) Dr. S. Simonian (NL) © CBG-MEB 1 Introduction • Problem statement: – What is the (un-)labelled use of individual macrolides in ocular, stomatologic, GI and SSTI infections? – What is the ultimate list of practical indications in these areas? • Approach: – Review information documented in data sheets & clinical literature. – From current global situation to agreed ----> by reconciliation of clinical experience with regulatory process----> state of the art indications. Focus will be primarily on systemic macrolides. © CBG-MEB 2 General Indications Ocular infections 1 ? Conjunctivitis (neonatal) C. trachomatis 2 ? Blepharitis Staphylococcal Stomatologic infections Eikenella corrodens? ? Periodontal disease3 Streptococci spp. Digestive tract infections HP peptic ulcer H. pylori* CJ Acute enteritis4 C. jejuni Cholecystitis5 SSTI Acne P. acnes Impetigo, erysipelas Streptococci spp. Furuncles, abcesses S. aureus Lyme disease?? B. burgdorferi Bartonella infections in HIV-patients ? Unlabelled use Questionable * In combination with PPI and other ?? antibiotics (eg. amoxicillin) As unlabelled alternative to beta-lactams and tetracyclines in early Lyme disease. As second-line in case tetracyline is inappropriate. © CBG-MEB 3 Indication status: Labelled(!), unlabelled & in research Ery Rox Clar Azit Telit Jos Spir Ocular infections Conjunctivitis(of the newborn) ! Blepharitis ! Trachoma -
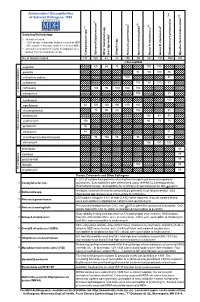
Antimicrobial Susceptibilities of Selected Pathogens, 1999
✔ Antimicrobial Susceptibilities * * † 7 of Selected Pathogens, 1999 8 e † a 2 ✔ ✔ † 3 ✔ † 4 5 6 culosis * r 1 urium spp. m L spp. Sampling Methodology 2 L † all isolates tested * ~ 20% sample of statewide isolates received at MDH spp. ~10% sample of statewide isolates received at MDH Salmonella ** all isolates tested from 7-county metropolitan area oup A streptococci ✔ oup B streptococci r isolates from a normally sterile site r Other (non-typhoidal) G Campylobacter Salmonella typhi Shigella Neisseria gonorrhoeae Neisseria meningitidis G Streptococcus pneumoni Mycobacterium tube No. of Isolates Tested 131 160 43 20 250 55 162 192 559 163 123456 123456% Susceptib123456le 123456123456 123456 123451623456 123451623456 ampicillin 1234561234566012345686 123456 15123456123456 100 100 123456123456 123451623451623451623451623456 123456 penicillin 123456123456123456123456123456 98100 100 76 123456 123456123456123456123456123456123456123456123456 123456 123451623451623451623456 123451623451623456 123456 cefuroxime sodium 123456123456123456123456 100123456123456123456 81 123456 123456123456123456123456123456123456123456123456 123456 cefotaxime 123451623451623451623451623456 100 100100 83 123456 123456123456123456123456123456 123456123456123456123456 123456 123456123456123456123456 ceftriaxone 123456 100 95 100 100 100 123451623451623451623456 -lactam antibiotics 123456123456123456123456123456 123456123456123456123456 β 123451623451623451623451623456 123451623456 123456 meropenem 123456123456123456123456123456 100 123456123456 83 123456 123456123456123456123456123456123456123456123456 -

Microbial NAD Metabolism: Lessons from Comparative Genomics
Dartmouth College Dartmouth Digital Commons Dartmouth Scholarship Faculty Work 9-2009 Microbial NAD Metabolism: Lessons from Comparative Genomics Francesca Gazzaniga Rebecca Stebbins Sheila Z. Chang Mark A. McPeek Dartmouth College Charles Brenner Carver College of Medicine Follow this and additional works at: https://digitalcommons.dartmouth.edu/facoa Part of the Biochemistry Commons, Genetics and Genomics Commons, Medicine and Health Sciences Commons, and the Microbiology Commons Dartmouth Digital Commons Citation Gazzaniga, Francesca; Stebbins, Rebecca; Chang, Sheila Z.; McPeek, Mark A.; and Brenner, Charles, "Microbial NAD Metabolism: Lessons from Comparative Genomics" (2009). Dartmouth Scholarship. 1191. https://digitalcommons.dartmouth.edu/facoa/1191 This Article is brought to you for free and open access by the Faculty Work at Dartmouth Digital Commons. It has been accepted for inclusion in Dartmouth Scholarship by an authorized administrator of Dartmouth Digital Commons. For more information, please contact [email protected]. MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS, Sept. 2009, p. 529–541 Vol. 73, No. 3 1092-2172/09/$08.00ϩ0 doi:10.1128/MMBR.00042-08 Copyright © 2009, American Society for Microbiology. All Rights Reserved. Microbial NAD Metabolism: Lessons from Comparative Genomics Francesca Gazzaniga,1,2 Rebecca Stebbins,1,2 Sheila Z. Chang,1,2 Mark A. McPeek,2 and Charles Brenner1,3* Departments of Genetics and Biochemistry and Norris Cotton Cancer Center, Dartmouth Medical School, Lebanon, New Hampshire -

Haemophilus Influenzae HP1 Bacteriophage Encodes a Lytic
International Journal of Molecular Sciences Article Haemophilus influenzae HP1 Bacteriophage Encodes a Lytic Cassette with a Pinholin and a Signal-Arrest-Release Endolysin Monika Adamczyk-Popławska * , Zuzanna Tracz-Gaszewska , Przemysław Lasota, Agnieszka Kwiatek and Andrzej Piekarowicz Department of Molecular Virology, Institute of Microbiology, Faculty of Biology, University of Warsaw, Miecznikowa 1, 02-096 Warsaw, Poland; [email protected] (Z.T.-G.); [email protected] (P.L.); [email protected] (A.K.); [email protected] (A.P.) * Correspondence: [email protected]; Tel.: +48-22-554-1419 Received: 9 May 2020; Accepted: 2 June 2020; Published: 4 June 2020 Abstract: HP1 is a temperate bacteriophage, belonging to the Myoviridae family and infecting Haemophilus influenzae Rd. By in silico analysis and molecular cloning, we characterized lys and hol gene products, present in the previously proposed lytic module of HP1 phage. The amino acid sequence of the lys gene product revealed the presence of signal-arrest-release (SAR) and muraminidase domains, characteristic for some endolysins. HP1 endolysin was able to induce lysis on its own when cloned and expressed in Escherichia coli, but the new phage release from infected H. influenzae cells was suppressed by inhibition of the secretion (sec) pathway. Protein encoded by hol gene is a transmembrane protein, with unusual C-out and N-in topology, when overexpressed/activated. Its overexpression in E. coli did not allow the formation of large pores (lack of leakage of β-galactosidase), but caused cell death (decrease in viable cell count) without lysis (turbidity remained constant). These data suggest that lys gene encodes a SAR-endolysin and that the hol gene product is a pinholin. -

Haemophilus Influenzae Genome Evolution During Persistence in The
Haemophilus influenzae genome evolution during PNAS PLUS persistence in the human airways in chronic obstructive pulmonary disease Melinda M. Pettigrewa, Christian P. Ahearnb,c, Janneane F. Gentd, Yong Konge,f,g, Mary C. Gallob,c, James B. Munroh,i, Adonis D’Melloh,i, Sanjay Sethic,j,k, Hervé Tettelinh,i,1, and Timothy F. Murphyb,c,l,1,2 aDepartment of Epidemiology of Microbial Diseases, Yale School of Public Health, New Haven, CT 06510; bDepartment of Microbiology and Immunology, University at Buffalo, The State University of New York, Buffalo, NY 14203; cClinical and Translational Research Center, University at Buffalo, The State University of New York, Buffalo, NY 14203; dDepartment of Environmental Health Sciences, Yale School of Public Health, New Haven, CT 06510; eDepartment of Biostatistics, Yale School of Public Health, New Haven, CT 06510; fDepartment of Molecular Biophysics and Biochemistry, Yale School of Medicine, New Haven, CT 06510; gW.M. Keck Foundation Biotechnology Resource Laboratory, Yale School of Medicine, New Haven, CT 06510; hInstitute for Genome Sciences, University of Maryland School of Medicine, Baltimore, MD 21201; iDepartment of Microbiology and Immunology, University of Maryland School of Medicine, Baltimore, MD 21201; jDivision of Pulmonary, Critical Care and Sleep Medicine, Department of Medicine, University at Buffalo, The State University of New York, Buffalo, NY 14203; kDepartment of Medicine, Veterans Affairs Western New York Healthcare System, Buffalo, NY 14215; and lDivision of Infectious Diseases, Department of Medicine, University at Buffalo, The State University of New York, Buffalo, NY 14203 Edited by Rino Rappuoli, GSK Vaccines, Siena, Italy, and approved February 27, 2018 (received for review November 10, 2017) Nontypeable Haemophilus influenzae (NTHi) exclusively colonize and adaptation cannot be accurately studied in vitro or in animal infect humans and are critical to the pathogenesis of chronic obstruc- models as a result of the unique physiological and immunological tive pulmonary disease (COPD). -

Lung Microbiome Dynamics in Chronic Obstructive Pulmonary Disease Exacerbations
ERJ Express. Published on February 25, 2016 as doi: 10.1183/13993003.01406-2015 ORIGINAL ARTICLE IN PRESS | CORRECTED PROOF Lung microbiome dynamics in chronic obstructive pulmonary disease exacerbations Zhang Wang1,7, Mona Bafadhel2,7, Koirobi Haldar3,6, Aaron Spivak1, David Mayhew1, Bruce E. Miller4, Ruth Tal-Singer4, Sebastian L. Johnston5, Mohammadali Yavari Ramsheh3, Michael R. Barer3, Christopher E. Brightling3,6,8 and James R. Brown1,8 Affiliations: 1Computational Biology, Target Sciences, GSK R&D, Collegeville, PA, USA. 2Respiratory Medicine Unit, Nuffield Department of Clinical Medicine, University of Oxford, Oxford, UK. 3Institute for Lung Health, National Institute for Health Research Respiratory Biomedical Research Unit, Department of Infection, Immunity and Inflammation, University of Leicester, Leicester, UK. 4Respiratory Therapy Area Unit, GSK R&D, King of Prussia, PA, USA. 5Airway Disease Infection Section, National Heart and Lung Institute, Imperial College London, London, UK. 6Department of Health Sciences, University of Leicester, Leicester, UK. 7These authors contributed equally. 8Both authors contributed equally. Correspondence: Christopher E. Brightling, Institute for Lung Health, Clinical Sciences Wing, University Hospitals of Leicester, Leicester, LE3 9QP, UK. E-mail: [email protected] ABSTRACT Increasing evidence suggests that the lung microbiome plays an important role in chronic obstructive pulmonary disease (COPD) severity. However, the dynamics of the lung microbiome during COPD exacerbations and its potential role in disease aetiology remain poorly understood. We completed a longitudinal 16S ribosomal RNA survey of the lung microbiome on 476 sputum samples collected from 87 subjects with COPD at four visits defined as stable state, exacerbation, 2 weeks post-therapy and 6 weeks recovery.