AN ABSTRACT of the DISSERTATION of Veli Erdogan
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

An Abstract of the Thesis Of
AN ABSTRACT OF THE THESIS OF Annie M. Chozinski for the degree of Master of Science in Horticulture presented on November 23. 1994. Title: The Evaluation of Cold Hardiness in Corvlus. Abstract approved: Shawn A. Mehlenbacher Anthesis of both staminate and pistillate flowers of Cory/us occurs in midwinter. To insure adequate pollination and nut set, these flowers must attain a sufficient hardiness level to withstand low temperatures. This study estimated cold hardiness of Cory/us cultivars and species using laboratory freezing of shoots without artificial hardening. In December, January and February of 1991-92 and 1992-93, one-year stems were collected 0 0 and frozen at regular intervals from -10 C to -38 C/ and visual browning assessed survival approximately 10 days after freezing. Elongated catkins were clipped prior to freezing. Percent flower bud survival was calculated and plotted against temperature. Linear regression generated an equation relating percent bud survival to temperature. From this equation, estimates of the LT^ (lethal temperature for 50% of the buds) was calculated for catkins, female inflorescences, and vegetative buds. C. avellana L. catkins, on average, were less hardy in both December and January than female inflorescences and vegetative buds. Maximum hardiness was reached in December and nearly all had elongated prior to the February freeze. Cultivars with the most hardy catkins were 'Morell', 'Brixnut', 'Creswell', 'Gem', 'Giresun OSU 54.080', 'Hall's Giant', 'Riccia di Talanico', 'Montebello' and 'Rode Zeller'. Maximum hardiness was observed for both vegetative and pistillate buds in January and was followed by a marked loss of hardiness in February. -
Qrno. 1 2 3 4 5 6 7 1 CP 2903 77 100 0 Cfcl3
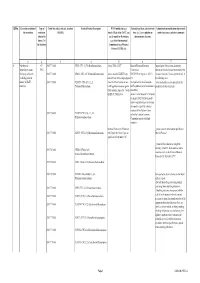
QRNo. General description of Type of Tariff line code(s) affected, based on Detailed Product Description WTO Justification (e.g. National legal basis and entry into Administration, modification of previously the restriction restriction HS(2012) Article XX(g) of the GATT, etc.) force (i.e. Law, regulation or notified measures, and other comments (Symbol in and Grounds for Restriction, administrative decision) Annex 2 of e.g., Other International the Decision) Commitments (e.g. Montreal Protocol, CITES, etc) 12 3 4 5 6 7 1 Prohibition to CP 2903 77 100 0 CFCl3 (CFC-11) Trichlorofluoromethane Article XX(h) GATT Board of Eurasian Economic Import/export of these ozone destroying import/export ozone CP-X Commission substances from/to the customs territory of the destroying substances 2903 77 200 0 CF2Cl2 (CFC-12) Dichlorodifluoromethane Article 46 of the EAEU Treaty DECISION on August 16, 2012 N Eurasian Economic Union is permitted only in (excluding goods in dated 29 may 2014 and paragraphs 134 the following cases: transit) (all EAEU 2903 77 300 0 C2F3Cl3 (CFC-113) 1,1,2- 4 and 37 of the Protocol on non- On legal acts in the field of non- _to be used solely as a raw material for the countries) Trichlorotrifluoroethane tariff regulation measures against tariff regulation (as last amended at 2 production of other chemicals; third countries Annex No. 7 to the June 2016) EAEU of 29 May 2014 Annex 1 to the Decision N 134 dated 16 August 2012 Unit list of goods subject to prohibitions or restrictions on import or export by countries- members of the -

Filberts PART I
Station Bulletin 208 August, 1924 Oregon Agricultural College Experiment Station Filberts PART I. GROWING FILBERTS IN OREGON PART II. EXPERIMENTAL DATA ON FILBERT POLLINATION By C. E. SCHUSTER CORVALLIS, OREGON The regular bulletins of the Station are sent free to the residents of Oregon who request them. BOARD OF REGENTS OF THE OREGON AGRICULTURAL COLLEGE AND EXPERIMENT STATION HON.J. K. WEATI-IERFORS, President Albany I-ION. JEFEERSON MYERS, Secretary Portland Hon. B. F. IRVINE, Treasurer Portland HON.WALTER M. PIERCE, Governor Salem - HON.SAss A. KOZER, Secretary of State--.. - Salem I-ION. J A. CHURCHILL, Superintendent of Public instruction ......Salem HON. GEommcE A. PALM ITER, Master of State Grange - Hood River - HON. K. B. ALDRICH . - . .Pertdleton Hon. SAM H. BROSvN Gervams HON.HARRY BAILEY .......... Lakeview I-ION. Geo. M. CORNWALL Portland Hon. M. S. \OODCOCX Corvallis Hon. E. E. \VILON Corvallis STATION STAFF 'N. J. KERR, D.Sc., LL.D... President J. T. JARDINE, XIS Director E. T. DEED, B.S., AD... Editor H. P. BARSS, A.B., S.M - - .Plant Pathologist B. B. BAYLES - Jr Plant Breeder, Office of Cer. loses., U. S. Dept. of Agri. P. M.BRAND-C,B.S , A M Dairy Husbandmami - Horticulturist (Vegetable Gardening) A.G. G. C. BOUQUET, BROWN, B.S B.......Horticulturist, Hood River Br Exp. Station, Hood River V. S. BROWN, AD., M S Horticulturist in Charge D. K. BULL1S, B.S - Assistant Chemist LEROY CHILOC, AD - .Supt Hood River Branch Exp. Station, Hood River V. Corson, MS. .. Bacteriologist K. DEAN, B.S..............Supt. Umatilla Brsnch Exp. Station, Hermiston FLOYD M. -

Filbert European Corylus Avellana Corylus Avellana Commonly Called
Filbert European Corylus avellana Corylus avellana commonly called European Filbert, European hazel, cobnut and Harry Lauder’s walking stick is a deciduous, thicket-forming, multi-trunked suckering shrub. Common names of filbert and hazel are likely interchangeable. Hazel is more often used in reference to wild specimens and filbert is more likely to be used in reference to cultivated plants. The filbert nuts to be produced in commerce primarily come from plants (C. avellano x C. maxima). ‘Contorta’, commonly called contorted filbert, corkscrew hazel or Harry Lauder’s Walking Stick, is contorted version of the species plant. It was discovered growing as a sport in an English hedgerow In the mid-1800s by Victorian Gardner Cannon Ellacombe. This plant was given the common name of Harry Lauder’s walking stick in the 1900s in honor of the Scottish entertainer Harry Lauder. The European Filbert leaves are dark green, slightly covered with fine soft hairs above and beneath; alternate; 2-4” in length, somewhat circular to egg – shaped or heart – shaped, abruptly tapers to a point at apex, edge doubly toothed, often with lobes, petiole ¼” to ½” long. The twigs are brown, glandular – hairy. Buds green to brown, hairless with hairy scale; overlapping, egg shaped to round. Flowers/Fruit: Flowers monoecious; male flowers are large (2”to 3”) catkins, yellow – brown, late winter to early spring blooming; female flowers inconspicuous. Fruit a nut; nuts inside involucre, which is toothed or lubed and nearly the length of the nut; ¾” in length; edible fruit grown commercially as a crop. European Filbert bark is pale to gray – brown, smoother with age, not an ornamental feature. -

Nutritive Value and Degradability of Leaves from Temperate Woody Resources for Feeding Ruminants in Summer
3rd European Agroforestry Conference Montpellier, 23-25 May 2016 Silvopastoralism (poster) NUTRITIVE VALUE AND DEGRADABILITY OF LEAVES FROM TEMPERATE WOODY RESOURCES FOR FEEDING RUMINANTS IN SUMMER Emile JC 1*, Delagarde R 2, Barre P 3, Novak S 1 Corresponding author: [email protected] mailto:(1) INRA, UE 1373 FERLUS, 86600 Lusignan, France (2) INRA, UMR 1348 INRA-Agrocampus Ouest, 35590 Saint-Gilles, France (3) INRA, UR 4 URP3F, 86600 Lusignan, France 1/ Introduction Integrating agroforestry in livestock farming systems may be a real opportunity in the current climatic, social and economic conditions. Trees can contribute to improve welfare of grazing ruminants. The production of leaves from woody plants may also constitute a forage resource for livestock (Papanastasis et al. 2008) during periods of low grasslands production (summer and autumn). To know the potential of leaves from woody plants to be fed by ruminants, including dairy females, the nutritive value of these new forages has to be evaluated. References on nutritive values that already exist for woody plants come mainly from tropical or Mediterranean climatic conditions (http://www.feedipedia.org/) and very few data are currently available for the temperate regions. In the frame of a long term mixed crop-dairy system experiment integrating agroforestry (Novak et al. 2016), a large evaluation of leaves from woody resources has been initiated. The objective of this evaluation is to characterise leaves of woody forage resources potentially available for ruminants (hedgerows, coppices, shrubs, pollarded trees), either directly by browsing or fed after cutting. This paper presents the evaluation of a first set of 12 woody resources for which the feeding value is evaluated through their protein and fibre concentrations, in vitro digestibility (enzymatic method) and effective ruminal degradability. -

Descriptors for Hazelnut (Corylus Avellana L.)
Descriptors for Hazelnut(Corylus avellana L.) List of Descriptors Allium (E, S) 2001 Pearl millet (E/F) 1993 Almond (revised)* (E) 1985 Pepino (E) 2004 Apple* (E) 1982 Phaseolus acutifolius (E) 1985 Apricot* (E) 1984 Phaseolus coccineus* (E) 1983 Avocado (E/S) 1995 Phaseolus lunatus (P) 2001 Bambara groundnut (E, F) 2000 Phaseolus vulgaris* (E, P) 1982 Banana (E, S, F) 1996 Pigeonpea (E) 1993 Barley (E) 1994 Pineapple (E) 1991 Beta (E) 1991 Pistachio (A, R, E, F) 1997 Black pepper (E/S) 1995 Pistacia (excluding Pistacia vera) (E) 1998 Brassica and Raphanus (E) 1990 Plum* (E) 1985 Brassica campestris L. (E) 1987 Potato variety* (E) 1985 Buckwheat (E) 1994 Quinua* (E) 1981 Cañahua (S) 2005 Rambutan 2003 Capsicum (E/S) 1995 Rice* (E) 2007 Cardamom (E) 1994 Rocket (E, I) 1999 Carrot (E, S, F) 1998 Rye and Triticale* (E) 1985 Cashew* (E) 1986 Safflower* (E) 1983 Cherry* (E) 1985 Sesame (E) 2004 Chickpea (E) 1993 Setaria italica and S. pumilia (E) 1985 Citrus (E, F, S) 1999 Shea tree (E) 2006 Coconut (E) 1995 Sorghum (E/F) 1993 Coffee (E, S, F) 1996 Soyabean* (E/C) 1984 Cotton (revised)* (E) 1985 Strawberry (E) 1986 Cowpea (E, P)* 1983 Sunflower* (E) 1985 Cultivated potato* (E) 1977 Sweet potato (E/S/F) 1991 Date Palm (F) 2005 Taro (E, F, S) 1999 Durian (E) 2007 Tea (E, S, F) 1997 Echinochloa millet* (E) 1983 Tomato (E, S, F) 1996 Eggplant (E/F) 1990 Tropical fruit (revised)* (E) 1980 Faba bean* (E) 1985 Ulluco (S) 2003 Fig (E) 2003 Vigna aconitifolia and V. -

Evidence of Low Chloroplast Genetic Diversity in Two Carpinus Species
Available online: www.notulaebotanicae.ro Print ISSN 0255-965X; Electronic 1842-4309 Not Bot Horti Agrobo, 2017, 45(1):316-322. DOI:10.15835/nbha45110799 Original Article Evidence of Low Chloroplast Genetic Diversity in Two Carpinus Species in the Northern Balkans Mihaela Cristina CĂRĂBUŞ 1, Alexandru Lucian CURTU 1, Dragoş POSTOLACHE 2, Elena CIOCÎRLAN 1, Neculae ŞOFLETEA 1* 1Transilvania University of Brasov, Department of Forest Sciences, 1 Şirul Beethoven, 500123, Braşov, Romania; [email protected] ; [email protected] ; [email protected] ; [email protected] (*corresponding author) 2National Research and Development Institute in Forestry “Marin Dracea”, 65 Horea Str., 400275 Cluj-Napoca, Romania; [email protected] Abstract Genetic diversity and differentiation in two Carpinus species ( C. betulus and C. orientalis ) occurring in Romania was investigated by using three chloroplast Simple Sequence Repeat markers (cpSSRs). A total of 96 and 32 individuals were sampled in eighteen C. betulus and six C. orientalis populations, respectively. A total of four chloroplast haplotypes were observed. Two haplotypes were specific for C. betulus and two for C. orientalis . Most of C. betulus populations were fixed for the predominant haplotype (H1), which was observed in 82% of the individuals. All C. orientalis populations were fixed for one haplotype or the other. Populations with haplotype (H3) are spread in southern Romania and the haplotype (H4) was observed at the northern limit of C. orientalis natural distribution range. Genetic differentiation among populations was moderate in C. betulus (GST = 0.422), compared to the high value observed in C. orientalis (GST = 1.000), which can be explained by the occurrence of a distinct haplotype in the peripheral population. -

Effects of Processing Treatments on Nutritional Quality of Raw Almond (Terminalia Catappa Linn.) Kernels
Available online a t www.pelagiaresearchlibrary.com Pelagia Research Library Advances in Applied Science Research, 2016, 7(1):1-7 ISSN: 0976-8610 CODEN (USA): AASRFC Effects of processing treatments on nutritional quality of raw almond (Terminalia catappa Linn.) kernels *Makinde Folasade M. and Oladunni Subomi S. Department of Food Science and Technology, Bowen University, Iwo, Osun State, Nigeria _____________________________________________________________________________________________ ABSTRACT Almond (Terminalia catappa Linn) is one of the lesser utilized oil kernel distributed throughout the tropics including Nigeria ecosystem. In this research work, the effects of soaking, blanching, autoclaving and roasting on the proximate, mineral, vitamin and anti-nutritional concentrations of almond kernel were determined. The result of chemical composition revealed that raw almond kernel contained 11.93% moisture, 23.0% crude protein, 48.1% crude fat, 2.43% crude fiber, 2.69% ash, 12.0% carbohydrate, 0.35mg/100g thiamine, 0.15mg/100g riboflavin, 0.19mg/100g niacin and minerals among which the most important are potassium (9.87 mg/100g), calcium (4.66 mg/100g) and magnesium (4.45 mg/100g). Tannin, phytate and oxalate concentration in raw almond kernel were 0.15, 0.13 and 0.15mg/100g respectively. Increase in ash and fiber was noted for treated samples with time compared to raw almond. Compared to untreated kernels, soaking, blanching and autoclaving decreased fat content but there was increase during roasting of the kernels. Mineral concentrations were significantly increased by various treatments compared to raw kernel. However, roasting for 15 min resulted in highest increase in potassium (41.2 percent), calcium (45.1 percent), phosphorus (43.3 percent) and magnesium (43.6 percent). -

Hazelnuts Resistant to Eastern Filbert Blight: Are We There Yet?
Hazelnuts Resistant to Eastern Filbert Blight: Are We There Yet? Thomas J. Molnar, Ph.D. Plant Biology and Pathology Dept. Rutgers University The American Chestnut Society Annual Meeting October 22, 2011 Nut Tree Breeding at Rutgers Based on the tremendous genetic improvements demonstrated in several previously underutilized turf species, Dr. C. Reed Funk strongly believed similar work could be done with nut trees Title of project started in 1996: Underutilized Perennial Food Crops Genetic Improvement Tom Molnar and Reed Funk Program Adelphia Research Farm August 2001 Nut Breeding at Rutgers Starting in 1996, species of interest included – black walnuts, Persian walnuts and heartnuts – pecans, hickories Pecan shade trial – chestnuts, Adelphia 2000 – almonds, – hazelnuts We built a germplasm collection of over 25,000 trees planted across five Rutgers research farms – Cream Ridge Fruit Research Farm (Cream Ridge, NJ) – Adelphia Research Farm (Freehold, NJ) Pecan shade trial Adelphia 2008 – HF1, HF2, HF3 (North Brunswick, NJ) Nut Breeding at Rutgers Goals – Identify species that show the greatest potential for New Jersey and Mid- Atlantic region – Develop breeding program to create superior well- adapted cultivars that reliably produce high- quality, high-value crops • while requiring reduced inputs of pesticides, fungicides, management, etc. Nut Breeding at Rutgers While most species showed great promise for substantial improvement, we had to narrow our focus to be most effective Hazelnuts stood out as the species where we could make significant contributions in a relatively short period of time – Major focus since 2000 Hazelnuts at Rutgers Why we chose to focus on hazelnuts: – success of initial plantings made in 1996/1997 with few pests and diseases – short generation time and small plant size (4 years from seed to seed) – wide genetic diversity and the ability to hybridize different species – ease of making controlled crosses – backlog of information and breeding advances – existing technologies and markets for nuts Hazelnuts: Corylus spp. -

CBD First National Report
FIRST NATIONAL REPORT OF THE REPUBLIC OF SERBIA TO THE UNITED NATIONS CONVENTION ON BIOLOGICAL DIVERSITY July 2010 ACRONYMS AND ABBREVIATIONS .................................................................................... 3 1. EXECUTIVE SUMMARY ........................................................................................... 4 2. INTRODUCTION ....................................................................................................... 5 2.1 Geographic Profile .......................................................................................... 5 2.2 Climate Profile ...................................................................................................... 5 2.3 Population Profile ................................................................................................. 7 2.4 Economic Profile .................................................................................................. 7 3 THE BIODIVERSITY OF SERBIA .............................................................................. 8 3.1 Overview......................................................................................................... 8 3.2 Ecosystem and Habitat Diversity .................................................................... 8 3.3 Species Diversity ............................................................................................ 9 3.4 Genetic Diversity ............................................................................................. 9 3.5 Protected Areas .............................................................................................10 -

Eastern Filbert Blight Kaitlin Morey Gold*, UW-Madison Plant Pathology
XHT1253 Provided to you by: Eastern Filbert Blight Kaitlin Morey Gold*, UW-Madison Plant Pathology What is Eastern filbert blight? Eastern filbert blight is a potentially serious fungal disease found throughout the United States, including Wisconsin. It affects only Corylus species, commonly known as hazelnuts or filberts. On hazelnuts native to Wisconsin such as American hazelnut (Corylus americana) and beaked hazelnut (Corylus cornuta), the disease causes little significant damage, but on the commonly grown European hazelnut (Corylus avellana), including Harry Lauder’s walking stick (Corylus avellana ‘Contorta’), the disease is lethal. Turkish filbert (Corylus colurna) also appears to be highly susceptible. What does Eastern filbert blight look like? Eastern filbert blight causes cankers (i.e., dead, collapsed areas of bark) on branches or main trunks. Easily visible within the cankers are black, football-shaped stromata (the reproductive structures of the causal fungus). The stromata often form in rows of two. Cankers first appear on new twigs and expand over time. American hazelnut trees/shrubs are able to live almost indefinitely with Eastern filbert blight, forming a small number of slowly-expanding cankers (if any cankers form at all) that lead to limited branch dieback. On European hazelnut however, cankers will expand anywhere from one inch to three feet in a year, and can eventually form long, deep gouges or grooves on severely affected trees/shrubs. Eastern filbert blight can cause small black European hazelnuts typically die due to cankers that form in rows, or deep gouges in the bark of severely infected trees/shrubs. girdling from Eastern filbert blight within five to 10 years. -

Arborerum Spring Planting Notes
ARNOLDIA A continuation of the BULLETIN OF POPULAR INFORMATION _ of the Arnold Arboretum, Harvard University VOLUME 16 .~Z.41~ 4, 1956 NUMBERS 4-5 ARBORETUM SPRING PLANTING NOTES planting at the Arnold Arboretum goes on year after year, unheralded Sand PRING unsung, yet many extremely interesting plants will eventually grow into the public’s notice. Young plants are necessarily pruned very heavily when they are transplanted to the grounds here, so that frequently several years elapse be- fore such plants eventually reach any real size and become noticeable to the horticultural-minded visitor. However, a close scrutiny of any year’s planting list will show many an old variety being replaced in young form (making it pos- sible to remove diseased or damaged specimens), and many new plants being placed that have never been in the Arboretum collections before. Some are prob- ably new to any American planting. Brief information concerning a few of the trees and shrubs planted out this spring will be of interest m this respect. It should be pointed out that to reach planting size, these plants ha~ e been growing in the nurseries of the Arnold Arboretum anywhere from one to sixteen years. Some must be tested for hardiness before they are planted in the perma- nent collections ; others must be checked when they flower to make certain they are named correctly. So, although they may be noted in the following pages as "new" to the permanent collections, they have all been growing in the Arbore- tum nurseries for several years. This is only part of a lengthy testing program for new plants which is going on continually, year after year.