Sparus Aurata) in Its Expanding Northerly Range
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

DEEP SEA LEBANON RESULTS of the 2016 EXPEDITION EXPLORING SUBMARINE CANYONS Towards Deep-Sea Conservation in Lebanon Project
DEEP SEA LEBANON RESULTS OF THE 2016 EXPEDITION EXPLORING SUBMARINE CANYONS Towards Deep-Sea Conservation in Lebanon Project March 2018 DEEP SEA LEBANON RESULTS OF THE 2016 EXPEDITION EXPLORING SUBMARINE CANYONS Towards Deep-Sea Conservation in Lebanon Project Citation: Aguilar, R., García, S., Perry, A.L., Alvarez, H., Blanco, J., Bitar, G. 2018. 2016 Deep-sea Lebanon Expedition: Exploring Submarine Canyons. Oceana, Madrid. 94 p. DOI: 10.31230/osf.io/34cb9 Based on an official request from Lebanon’s Ministry of Environment back in 2013, Oceana has planned and carried out an expedition to survey Lebanese deep-sea canyons and escarpments. Cover: Cerianthus membranaceus © OCEANA All photos are © OCEANA Index 06 Introduction 11 Methods 16 Results 44 Areas 12 Rov surveys 16 Habitat types 44 Tarablus/Batroun 14 Infaunal surveys 16 Coralligenous habitat 44 Jounieh 14 Oceanographic and rhodolith/maërl 45 St. George beds measurements 46 Beirut 19 Sandy bottoms 15 Data analyses 46 Sayniq 15 Collaborations 20 Sandy-muddy bottoms 20 Rocky bottoms 22 Canyon heads 22 Bathyal muds 24 Species 27 Fishes 29 Crustaceans 30 Echinoderms 31 Cnidarians 36 Sponges 38 Molluscs 40 Bryozoans 40 Brachiopods 42 Tunicates 42 Annelids 42 Foraminifera 42 Algae | Deep sea Lebanon OCEANA 47 Human 50 Discussion and 68 Annex 1 85 Annex 2 impacts conclusions 68 Table A1. List of 85 Methodology for 47 Marine litter 51 Main expedition species identified assesing relative 49 Fisheries findings 84 Table A2. List conservation interest of 49 Other observations 52 Key community of threatened types and their species identified survey areas ecological importanc 84 Figure A1. -

Updated Checklist of Marine Fishes (Chordata: Craniata) from Portugal and the Proposed Extension of the Portuguese Continental Shelf
European Journal of Taxonomy 73: 1-73 ISSN 2118-9773 http://dx.doi.org/10.5852/ejt.2014.73 www.europeanjournaloftaxonomy.eu 2014 · Carneiro M. et al. This work is licensed under a Creative Commons Attribution 3.0 License. Monograph urn:lsid:zoobank.org:pub:9A5F217D-8E7B-448A-9CAB-2CCC9CC6F857 Updated checklist of marine fishes (Chordata: Craniata) from Portugal and the proposed extension of the Portuguese continental shelf Miguel CARNEIRO1,5, Rogélia MARTINS2,6, Monica LANDI*,3,7 & Filipe O. COSTA4,8 1,2 DIV-RP (Modelling and Management Fishery Resources Division), Instituto Português do Mar e da Atmosfera, Av. Brasilia 1449-006 Lisboa, Portugal. E-mail: [email protected], [email protected] 3,4 CBMA (Centre of Molecular and Environmental Biology), Department of Biology, University of Minho, Campus de Gualtar, 4710-057 Braga, Portugal. E-mail: [email protected], [email protected] * corresponding author: [email protected] 5 urn:lsid:zoobank.org:author:90A98A50-327E-4648-9DCE-75709C7A2472 6 urn:lsid:zoobank.org:author:1EB6DE00-9E91-407C-B7C4-34F31F29FD88 7 urn:lsid:zoobank.org:author:6D3AC760-77F2-4CFA-B5C7-665CB07F4CEB 8 urn:lsid:zoobank.org:author:48E53CF3-71C8-403C-BECD-10B20B3C15B4 Abstract. The study of the Portuguese marine ichthyofauna has a long historical tradition, rooted back in the 18th Century. Here we present an annotated checklist of the marine fishes from Portuguese waters, including the area encompassed by the proposed extension of the Portuguese continental shelf and the Economic Exclusive Zone (EEZ). The list is based on historical literature records and taxon occurrence data obtained from natural history collections, together with new revisions and occurrences. -

Triple Barcoding for a Hyperparasite, Its Parasitic Host, and the Host Itself
Parasite 28, 49 (2021) Ó C. Bouguerche et al., published by EDP Sciences, 2021 https://doi.org/10.1051/parasite/2021044 Available online at: www.parasite-journal.org RESEARCH ARTICLE OPEN ACCESS Triple barcoding for a hyperparasite, its parasitic host, and the host itself: a study of Cyclocotyla bellones (Monogenea) on Ceratothoa parallela (Isopoda) on Boops boops (Teleostei) Chahinez Bouguerche1,4, Fadila Tazerouti1, Delphine Gey2,3, and Jean-Lou Justine4,* 1 Université des Sciences et de la Technologie Houari Boumediene, Faculté des Sciences Biologiques, Laboratoire de Biodiversité et Environnement : Interactions – Génomes, BP 32, El Alia, Bab Ezzouar, Algiers, Algeria 2 Service de Systématique moléculaire, UMS 2700 CNRS, Muséum National d’Histoire Naturelle, Sorbonne Universités, 43 rue Cuvier, CP 26, 75231 Paris Cedex 05, France 3 UMR7245 MCAM, Muséum National d’Histoire Naturelle, 43 rue Cuvier, CP 52, 75231 Paris Cedex 05, France 4 Institut Systématique Évolution Biodiversité (ISYEB), Muséum National d’Histoire Naturelle, CNRS, Sorbonne Université, EPHE, Université des Antilles, 43 rue Cuvier, CP 51, 75231 Paris Cedex 05, France Received 15 October 2020, Accepted 7 May 2021, Published online 7 June 2021 Abstract – Cyclocotyla bellones Otto, 1823 (Diclidophoridae) is a monogenean characterised by an exceptional way of life. It is a hyperparasite that attaches itself to the dorsal face of isopods, themselves parasites in the buccal cavity of fishes. In this study, Cy. bellones was found on Ceratothoa parallela (Otto, 1828), a cymothoid isopod parasite of the sparid fish Boops boops off Algeria in the Mediterranean Sea. We provide, for the first time, molecular barcoding information of a hyperparasitic monogenean, the parasitic crustacean host, and the fish host, with COI sequences. -
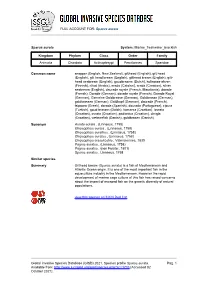
Sparus Aurata Global Invasive Species Database (GISD)
FULL ACCOUNT FOR: Sparus aurata Sparus aurata System: Marine_freshwater_brackish Kingdom Phylum Class Order Family Animalia Chordata Actinopterygii Perciformes Sparidae Common name snapper (English, New Zealand), gilthead (English), gilt head (English), gilt head bream (English), gilthead bream (English), gilt- head seabream (English), goudbrasem (Dutch), kultaotsa-ahven (Finnish), n'tad (Arabic), orada (Catalan), orada (Croatian), silver seabream (English), daurade royale (French, Mauritania), dorade (French), Dorade (German), dorade royale (French), Dorade Royal (German), Gemeine Goldbrasse (German), Goldbrasse (German), goldbrassen (German), Goldkopf (German), daurade (French), tsipoura (Greek), dorada (Spanish), dourada (Portuguese), cipura (Turkish), goud brasem (Dutch), komarca (Croatian), lovrata (Croatian), ovrata (Croatian), podlanica (Croatian), dinigla (Croatian), væbnerfisk (Danish), guldbrasen (Danish) Synonym Aurata aurata , (Linnaeus, 1758) Chrysophrys aurata , (Linnaeus, 1758) Chrysophrys aurathus , (Linnaeus, 1758) Chrysophrys auratus , (Linnaeus, 1758) Chrysophrys crassirostris , Valenciennes, 1830 Pagrus auratus , (Linnaeus, 1758) Pagrus auratus , (non Forster, 1801) Sparus auratus , Linnaeus, 1758 Similar species Summary Gilthead bream (Sparus aurata) is a fish of Mediterranean and Atlantic Ocean origin. It is one of the most important fish in the aquaculture industry in the Mediterranean. However the rapid development of marine cage culture of this fish has raised concerns about the impact of escaped fish on the genetic diversity of natural populations. view this species on IUCN Red List Global Invasive Species Database (GISD) 2021. Species profile Sparus aurata. Pag. 1 Available from: http://www.iucngisd.org/gisd/species.php?sc=1703 [Accessed 02 October 2021] FULL ACCOUNT FOR: Sparus aurata Species Description The gilthead bream is a Mediterranean fish reaching a maximum of 70 cm length and 6 kg in weight (Balart et al., 2009). -

Marine Fishes from Galicia (NW Spain): an Updated Checklist
1 2 Marine fishes from Galicia (NW Spain): an updated checklist 3 4 5 RAFAEL BAÑON1, DAVID VILLEGAS-RÍOS2, ALBERTO SERRANO3, 6 GONZALO MUCIENTES2,4 & JUAN CARLOS ARRONTE3 7 8 9 10 1 Servizo de Planificación, Dirección Xeral de Recursos Mariños, Consellería de Pesca 11 e Asuntos Marítimos, Rúa do Valiño 63-65, 15703 Santiago de Compostela, Spain. E- 12 mail: [email protected] 13 2 CSIC. Instituto de Investigaciones Marinas. Eduardo Cabello 6, 36208 Vigo 14 (Pontevedra), Spain. E-mail: [email protected] (D. V-R); [email protected] 15 (G.M.). 16 3 Instituto Español de Oceanografía, C.O. de Santander, Santander, Spain. E-mail: 17 [email protected] (A.S); [email protected] (J.-C. A). 18 4Centro Tecnológico del Mar, CETMAR. Eduardo Cabello s.n., 36208. Vigo 19 (Pontevedra), Spain. 20 21 Abstract 22 23 An annotated checklist of the marine fishes from Galician waters is presented. The list 24 is based on historical literature records and new revisions. The ichthyofauna list is 25 composed by 397 species very diversified in 2 superclass, 3 class, 35 orders, 139 1 1 families and 288 genus. The order Perciformes is the most diverse one with 37 families, 2 91 genus and 135 species. Gobiidae (19 species) and Sparidae (19 species) are the 3 richest families. Biogeographically, the Lusitanian group includes 203 species (51.1%), 4 followed by 149 species of the Atlantic (37.5%), then 28 of the Boreal (7.1%), and 17 5 of the African (4.3%) groups. We have recognized 41 new records, and 3 other records 6 have been identified as doubtful. -

Cymothoidae) from Sub-Sahara Africa
Biodiversity and systematics of branchial cavity inhabiting fish parasitic isopods (Cymothoidae) from sub-Sahara Africa S van der Wal orcid.org/0000-0002-7416-8777 Previous qualification (not compulsory) Dissertation submitted in fulfilment of the requirements for the Masters degree in Environmental Sciences at the North-West University Supervisor: Prof NJ Smit Co-supervisor: Dr KA Malherbe Graduation May 2018 23394536 TABLE OF CONTENTS LIST OF FIGURES ................................................................................................................... VI LIST OF TABLES .................................................................................................................. XIII ABBREVIATIONS .................................................................................................................. XIV ACKNOWLEDGEMENTS ....................................................................................................... XV ABSTRACT ........................................................................................................................... XVI CHAPTER 1: INTRODUCTION ................................................................................................. 1 1.1 Subphylum Crustacea Brünnich, 1772 ............................................................ 2 1.2 Order Isopoda Latreille, 1817 ........................................................................... 2 1.3 Parasitic Isopoda ............................................................................................. -

ASFIS ISSCAAP Fish List February 2007 Sorted on Scientific Name
ASFIS ISSCAAP Fish List Sorted on Scientific Name February 2007 Scientific name English Name French name Spanish Name Code Abalistes stellaris (Bloch & Schneider 1801) Starry triggerfish AJS Abbottina rivularis (Basilewsky 1855) Chinese false gudgeon ABB Ablabys binotatus (Peters 1855) Redskinfish ABW Ablennes hians (Valenciennes 1846) Flat needlefish Orphie plate Agujón sable BAF Aborichthys elongatus Hora 1921 ABE Abralia andamanika Goodrich 1898 BLK Abralia veranyi (Rüppell 1844) Verany's enope squid Encornet de Verany Enoploluria de Verany BLJ Abraliopsis pfefferi (Verany 1837) Pfeffer's enope squid Encornet de Pfeffer Enoploluria de Pfeffer BJF Abramis brama (Linnaeus 1758) Freshwater bream Brème d'eau douce Brema común FBM Abramis spp Freshwater breams nei Brèmes d'eau douce nca Bremas nep FBR Abramites eques (Steindachner 1878) ABQ Abudefduf luridus (Cuvier 1830) Canary damsel AUU Abudefduf saxatilis (Linnaeus 1758) Sergeant-major ABU Abyssobrotula galatheae Nielsen 1977 OAG Abyssocottus elochini Taliev 1955 AEZ Abythites lepidogenys (Smith & Radcliffe 1913) AHD Acanella spp Branched bamboo coral KQL Acanthacaris caeca (A. Milne Edwards 1881) Atlantic deep-sea lobster Langoustine arganelle Cigala de fondo NTK Acanthacaris tenuimana Bate 1888 Prickly deep-sea lobster Langoustine spinuleuse Cigala raspa NHI Acanthalburnus microlepis (De Filippi 1861) Blackbrow bleak AHL Acanthaphritis barbata (Okamura & Kishida 1963) NHT Acantharchus pomotis (Baird 1855) Mud sunfish AKP Acanthaxius caespitosa (Squires 1979) Deepwater mud lobster Langouste -

Field Guide to the Living Marine Resources of Namibia.Pdf
FAOSPECIESIDENTIFICATIONGUIDEFORFISHERYPURPOSES ISSN 1020-6868 FIELD GUIDE TO THE LIVING MARINE RESOURCES OF NAMIBIA Food and NORAD Agriculture Organization Norwegian of Agency for the International United Nations Development FAO SPECIES IDENTIFICATION FIELD GUIDE FOR FISHERY PURPOSES THE LIVING MARINE RESOURCES OF NAMIBIA by G. Bianchi Institute of Marine Research P.O. Box 1870, N-5024 Bergen, Norway K.E. Carpenter Department of Biological Sciences Old Dominion University Norfolk, Virginia 23529 USA J.-P. Roux Ministry of Fisheries and Marine Resources P.O. Box 394 Lüderitz, Namibia F.J. Molloy Biology Departmant Faculty of Science University of Namibia Private Bag 31 Windhoek, Namibia D. Boyer and H.J. Boyer Ministry of Fisheries and Marine Resources P.O. Box 912 Swakopmund, Namibia With the financial support of NORAD Norwegian Agency for International Development INDEX FOOD AND AGRICULTURAL ORGANIZATION OF THE UNITED NATIONS ROME, 1999 The designations employed and the presentation of material in this publication do not imply the expression of any opinion whatsoever on the part of the Food and Agricultural Organization of the United Nations concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. M-40 ISBN 92-5-104345-0 All rights reserved. No part of this publication may be reproduced, stored in a retrieval system, or transmitted in any form or by any means, electronic, mechanical, photocopying or otherwise, without the prior permission of the copyright owner. Applications for such permission, with a statement of the purpose and extent of the reproduction, should be addressed to the Director, Publications Division, Food and Agriculture Organiztion of the United Nations, Viale delle Terme di Caracalla, 00100 Rome, Italy. -

Sparidentex Hasta): a Comparison with Other Farmed Sparid Species
fishes Review Macronutrient Requirements of Silvery-Black Porgy (Sparidentex hasta): A Comparison with Other Farmed Sparid Species Mansour Torfi Mozanzadeh 1,*, Jasem G. Marammazi 1, Morteza Yaghoubi 1, Naser Agh 2, Esmaeil Pagheh 1 and Enric Gisbert 3 1 South Iranian Aquaculture Research Center, P.O. Box 669 Ahwaz, Iran; [email protected] (J.G.M.); [email protected] (M.Y.); [email protected] (E.P.) 2 Artemia and Aquatic Research Institute, Urmia University, 57135 Urmia, Iran; [email protected] 3 Institut de Recerca i Tecnologia Agroalimentaries, Centre de Sant Carles de la Rápita (IRTA-SCR), Unitat de Cultius Experimentals, 43540 Sant Carles de la Ràpita, Spain; [email protected] * Correspondence: mansour.torfi@gmail.com Academic Editor: Francisco J. Moyano Received: 31 January 2017; Accepted: 5 May 2017; Published: 13 May 2017 Abstract: Silvery-black porgy (Sparidentex hasta) is recognized as one of the most promising fish species for aquaculture diversification in the Persian Gulf and the Oman Sea regions. In this regard, S. hasta has received considerable attention, and nutritional studies focused on establishing the nutritional requirements for improving diet formulation have been conducted during recent years. Considering the results from different dose–response nutritional studies on macronutrient requirements conducted in this species, it can be concluded that diets containing ca. 48% crude protein, 15% crude lipid, 15% carbohydrates and 20 KJ g−1 gross energy are recommended for on-growing S. hasta juveniles. In addition, the optimum essential amino acid profile for this species (expressed as g 16 g N−1), should be approximately arginine 5.3, lysine 6.0, threonine 5.2, histidine 2.5, isoleucine 4.6, leucine 5.4, methionine + cysteine 4.0 (in a diet containing 0.6 cysteine), phenylalanine + tyrosine 5.6 (in a diet containing 1.9 tyrosine), tryptophan 1.0 and valine 4.6. -

A Survey of Marine Bony Fishes of the Gaza Strip, Palestine
The Islamic University of Gaza الجــــــــــامعة اﻹســــــــــﻻميـة بغــــــــــــــــــزة Deanship of Research and Graduate Studies عمادة البحث العممي والدراسات العميا Faculty of Science كــــميـــــــــــــــــــــــــــــــة العـمـــــــــــــــــــــــــــــــــــــوم Biological Sciences Master Program ماجـســــــــتيـر العمــــــــــــــوم الحـياتيــــــــــة )Zoology) عمـــــــــــــــــــــــــــــــــــــــم حيـــــــــــــــــــــــــــــــــــــــوا A Survey of Marine Bony Fishes of the Gaza Strip, Palestine ِـخ ٌﻷؿّان اؼٌظٍّح اٌثذغٌح فً لطاع غؼج، فٍـطٍٓ By Huda E. Abu Amra B.Sc. Biology Supervised by Dr. Abdel Fattah N. Abd Rabou Associate Professor of Environmental Sciences A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science in Biological Science (Zoology) August, 2018 إلــــــــــــــغاع أٔا اٌّىلغ أصٔاٖ ِمضَ اٌغؿاٌح اٌتً تذًّ اؼٌٕىاْ: A Survey of Marine Bony Fishes in the Gaza Strip, Palestine ِـخ ٌﻷؿّان اؼٌظٍّح اٌثذغٌح فً لطاع غؼج، فٍـطٍٓ أقش تأٌ يا اشرًهد عهّٛ ْزِ انشعانح إًَا ْٕ َراض جٓذ٘ انخاص، تاعرصُاء يا ذًد اﻹشاسج إنّٛ حٛصًا ٔسد، ٔأٌ ْزِ انشعانح ككم أٔ أ٘ جضء يُٓا نى ٚقذو يٍ قثم اٜخشٍٚ نُٛم دسجح أٔ نقة عهًٙ أٔ تحصٙ نذٖ أ٘ يؤعغح ذعهًٛٛح أٔ تحصٛح أخشٖ. Declaration I understand the nature of plagiarism, and I am aware of the University’s policy on this. The work provided in this thesis, unless otherwise referenced, is the researcher's own work, and has not been submitted by others elsewhere for any other degree or qualification. اعى انطانثح ْذٖ عٛذ عٛذ أتٕ عًشج :Student's name انرٕقٛع Signature: Huda انراسٚخ Date: 8-8-2018 I ٔتٍجح اٌذىُ ػٍى أطغودح ِاجـتٍغ II Abstract The East Mediterranean Sea is home to a wealth of marine resources including the bony fishes (Class Osteichthyes), which constitute a capital source of protein worldwide. -

The Sparid Fishes of Pakistan, with New Distribution Records
Zootaxa 3857 (1): 071–100 ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ Article ZOOTAXA Copyright © 2014 Magnolia Press ISSN 1175-5334 (online edition) http://dx.doi.org/10.11646/zootaxa.3857.1.3 http://zoobank.org/urn:lsid:zoobank.org:pub:A26948F7-39C6-4858-B7FD-380E12F9BD34 The sparid fishes of Pakistan, with new distribution records PIRZADA JAMAL SIDDIQUI1, SHABIR ALI AMIR1, 2 & RAFAQAT MASROOR2 1Centre of Excellence in Marine Biology, University of Karachi, Karachi, Pakistan 2Pakistan Museum of Natural History, Garden Avenue, Shakarparian, Islamabad–44000, Pakistan Corresponding author Shabir Ali Amir, email: [email protected] Abstract The family sparidae is represented in Pakistan by 14 species belonging to eight genera: the genus Acanthopagrus with four species, A. berda, A. arabicus, A. sheim, and A. catenula; Rhabdosargus, Sparidentex and Diplodus are each represented by two species, R. sarba and R. haffara, Sparidentex hasta and S. jamalensis, and Diplodus capensis and D. omanensis, and the remaining four genera are represented by single species, Crenidens indicus, Argyrops spinifer, Pagellus affinis, and Cheimerius nufar. Five species, Acanthopagrus arabicus, A. sheim, A. catenula, Diplodus capensis and Rhabdosargus haffara are reported for the first time from Pakistani coastal waters. The Arabian Yellowfin Seabream Acanthopagrus ar- abicus and Spotted Yellowfin Seabream Acanthopagrus sheim have only recently been described from Pakistani waters, while Diplodus omanensis and Pagellus affinis are newly identified from Pakistan. Acanthopagrus catenula has long been incorrectly identified as A. bifasciatus, a species which has not been recorded from Pakistan. All species are briefly de- scribed and a key is provided for them. Key words: Acanthopagrus, Rhabdosargus, Sparidentex, Diplodus, Crenidens, Argyrops, Pagellus, Cheimerius, Spari- dae, Karachi, Pakistan Introduction The fishes belonging to family Sparidae, commonly known as porgies and seabreams, are widely distributed in tropical to temperate seas (Froese & Pauly, 2013). -
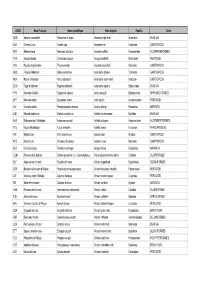
Liste Espèces
CODE Nom Français Nom scientifique Nom Anglais Famille Ordre ODQ Anomie cascabelle Pododesmus cepio Abalone jingle shell Anomiidae BIVALVIA ABX Ormeaux nca Haliotis spp Abalones nei Haliotidae GASTROPODA REN Sébaste rose Sebastes fasciatus Acadian redfish Scorpaenidae SCORPAENIFORMES YNA Acoupa toeroe Cynoscion acoupa Acoupa weakfish Sciaenidae PERCOIDEI HSV Pourpre aiguillonnee Thais aculeata Aculeate rock shell Muricidae GASTROPODA GBQ Troque d'Adanson Gibbula adansoni Adanson's gibbula Trochidae GASTROPODA NKA Natice d'Adanson Natica adansoni Adanson's moon snail Naticidae GASTROPODA GLW Tagal d'Adanson Tagelus adansonii Adanson's tagelus Solecurtidae BIVALVIA PYD Manchot d'Adélie Pygoscelis adeliae Adelie penguin Spheniscidae SPHENISCIFORMES QFT Maconde aden Synagrops adeni Aden splitfin Acropomatidae PERCOIDEI NIV Crevette adonis Parapenaeopsis venusta Adonis shrimp Penaeidae NATANTIA DJD Modiole adriatique Modiolus adriaticus Adriatic horse mussel Mytilidae BIVALVIA AAA Esturgeon de l'Adriatique Acipenser naccarii Adriatic sturgeon Acipenseridae ACIPENSERIFORMES FCV Fucus d'Adriatique Fucus virsoides Adriatic wrack Fucaceae PHAEOPHYCEAE IRR Mitre brûlée Mitra eremitarum Adusta miter Mitridae GASTROPODA KCE Murex bruni Chicoreus brunneus Adusta murex Muricidae GASTROPODA AES Crevette ésope Pandalus montagui Aesop shrimp Pandalidae NATANTIA CGM Poisson-chat, hybride Clarias gariepinus x C. macrocephalus Africa-bighead catfish, hybrid Clariidae SILURIFORMES SUF Ange de mer africain Squatina africana African angelshark Squatinidae SQUALIFORMES