„Automating the Analysis of Territory Mapping Data in Bird Monitoring“
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Dieter Thomas Tietze Editor How They Arise, Modify and Vanish
Fascinating Life Sciences Dieter Thomas Tietze Editor Bird Species How They Arise, Modify and Vanish Fascinating Life Sciences This interdisciplinary series brings together the most essential and captivating topics in the life sciences. They range from the plant sciences to zoology, from the microbiome to macrobiome, and from basic biology to biotechnology. The series not only highlights fascinating research; it also discusses major challenges associated with the life sciences and related disciplines and outlines future research directions. Individual volumes provide in-depth information, are richly illustrated with photographs, illustrations, and maps, and feature suggestions for further reading or glossaries where appropriate. Interested researchers in all areas of the life sciences, as well as biology enthusiasts, will find the series’ interdisciplinary focus and highly readable volumes especially appealing. More information about this series at http://www.springer.com/series/15408 Dieter Thomas Tietze Editor Bird Species How They Arise, Modify and Vanish Editor Dieter Thomas Tietze Natural History Museum Basel Basel, Switzerland ISSN 2509-6745 ISSN 2509-6753 (electronic) Fascinating Life Sciences ISBN 978-3-319-91688-0 ISBN 978-3-319-91689-7 (eBook) https://doi.org/10.1007/978-3-319-91689-7 Library of Congress Control Number: 2018948152 © The Editor(s) (if applicable) and The Author(s) 2018. This book is an open access publication. Open Access This book is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made. -

Bird Checklists of the World Country Or Region: Ghana
Avibase Page 1of 24 Col Location Date Start time Duration Distance Avibase - Bird Checklists of the World 1 Country or region: Ghana 2 Number of species: 773 3 Number of endemics: 0 4 Number of breeding endemics: 0 5 Number of globally threatened species: 26 6 Number of extinct species: 0 7 Number of introduced species: 1 8 Date last reviewed: 2019-11-10 9 10 Recommended citation: Lepage, D. 2021. Checklist of the birds of Ghana. Avibase, the world bird database. Retrieved from .https://avibase.bsc-eoc.org/checklist.jsp?lang=EN®ion=gh [26/09/2021]. Make your observations count! Submit your data to ebird. -

Progress in the Development of an Eurasian-African Bird Migration Atlas
CONVENTION ON UNEP/CMS/COP13/Inf.20 MIGRATORY 10 February 2020 SPECIES Original: English 13th MEETING OF THE CONFERENCE OF THE PARTIES Gandhinagar, India, 17 - 22 February 2020 Agenda Item 25 PROGRESS IN THE DEVELOPMENT OF AN EURASIAN-AFRICAN BIRD MIGRATION ATLAS (Submitted by the European Union of Bird Ringing (EURING) and the Institute of Avian Research) Summary: The African-Eurasian Bird Migration Atlas is being developed under the auspices of CMS in the framework of a Global Animal Migration Atlas, of which it constitutes a module. The African-Eurasian Bird Migration Atlas is being developed and compiled by the European Union of Bird Ringing (EURING) under a Project Cooperation Agreement (PCA) between the CMS Secretariat and the Institute of Avian Research, acting on behalf of EURING. The development of the African-Eurasian Bird Migration Atlas is funded with the contribution granted by the Government of Italy under the Migratory Species Champion Programme. This information document includes a progress report on the development of the various components of the project. The project is expected to be completed in 2021. UNEP/CMS/COP13/Inf.20 Eurasian-African Bird Migration Atlas progress report February 2020 Stephen Baillie1, Franz Bairlein2, Wolfgang Fiedler3, Fernando Spina4, Kasper Thorup5, Sam Franks1, Dorian Moss1, Justin Walker1, Daniel Higgins1, Roberto Ambrosini6, Niccolò Fattorini6, Juan Arizaga7, Maite Laso7, Frédéric Jiguet8, Boris Nikolov9, Henk van der Jeugd10, Andy Musgrove1, Mark Hammond1 and William Skellorn1. A report to the Convention on Migratory Species from the European Union for Bird Ringing (EURING) and the Institite of Avian Research, Wilhelmshaven, Germany 1. British Trust for Ornithology, Thetford, IP24 2PU, UK 2. -
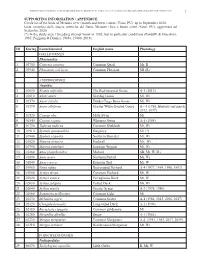
1 ID Euring Latin Binomial English Name Phenology Galliformes
BIRDS OF METAURO RIVER: A GREAT ORNITHOLOGICAL DIVERSITY IN A SMALL ITALIAN URBANIZING BIOTOPE, REQUIRING GREATER PROTECTION 1 SUPPORTING INFORMATION / APPENDICE Check list of the birds of Metauro river (mouth and lower course / Fano, PU), up to September 2020. Lista completa delle specie ornitiche del fiume Metauro (foce e basso corso /Fano, PU), aggiornata ad Settembre 2020. (*) In the study area 1 breeding attempt know in 1985, but in particolar conditions (Pandolfi & Giacchini, 1985; Poggiani & Dionisi, 1988a, 1988b, 2019). ID Euring Latin binomial English name Phenology GALLIFORMES Phasianidae 1 03700 Coturnix coturnix Common Quail Mr, B 2 03940 Phasianus colchicus Common Pheasant SB (R) ANSERIFORMES Anatidae 3 01690 Branta ruficollis The Red-breasted Goose A-1 (2012) 4 01610 Anser anser Greylag Goose Mi, Wi 5 01570 Anser fabalis Tundra/Taiga Bean Goose Mi, Wi 6 01590 Anser albifrons Greater White-fronted Goose A – 4 (1986, february and march 2012, 2017) 7 01520 Cygnus olor Mute Swan Mi 8 01540 Cygnus cygnus Whooper Swan A-1 (1984) 9 01730 Tadorna tadorna Common Shelduck Mr, Wi 10 01910 Spatula querquedula Garganey Mr (*) 11 01940 Spatula clypeata Northern Shoveler Mr, Wi 12 01820 Mareca strepera Gadwall Mr, Wi 13 01790 Mareca penelope Eurasian Wigeon Mr, Wi 14 01860 Anas platyrhynchos Mallard SB, Mr, W (R) 15 01890 Anas acuta Northern Pintail Mi, Wi 16 01840 Anas crecca Eurasian Teal Mr, W 17 01960 Netta rufina Red-crested Pochard A-4 (1977, 1994, 1996, 1997) 18 01980 Aythya ferina Common Pochard Mr, W 19 02020 Aythya nyroca Ferruginous -
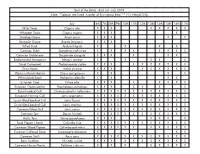
Best of the Baltic - Bird List - July 2019 Note: *Species Are Listed in Order of First Seeing Them ** H = Heard Only
Best of the Baltic - Bird List - July 2019 Note: *Species are listed in order of first seeing them ** H = Heard Only July 6th 7th 8th 9th 10th 11th 12th 13th 14th 15th 16th 17th Mute Swan Cygnus olor X X X X X X X X Whopper Swan Cygnus cygnus X X X X Greylag Goose Anser anser X X X X X Barnacle Goose Branta leucopsis X X X Tufted Duck Aythya fuligula X X X X Common Eider Somateria mollissima X X X X X X X X Common Goldeneye Bucephala clangula X X X X X X Red-breasted Merganser Mergus serrator X X X X X Great Cormorant Phalacrocorax carbo X X X X X X X X X X Grey Heron Ardea cinerea X X X X X X X X X Western Marsh Harrier Circus aeruginosus X X X X White-tailed Eagle Haliaeetus albicilla X X X X Eurasian Coot Fulica atra X X X X X X X X Eurasian Oystercatcher Haematopus ostralegus X X X X X X X Black-headed Gull Chroicocephalus ridibundus X X X X X X X X X X X X European Herring Gull Larus argentatus X X X X X X X X X X X X Lesser Black-backed Gull Larus fuscus X X X X X X X X X X X X Great Black-backed Gull Larus marinus X X X X X X X X X X X X Common/Mew Gull Larus canus X X X X X X X X X X X X Common Tern Sterna hirundo X X X X X X X X X X X X Arctic Tern Sterna paradisaea X X X X X X X Feral Pigeon ( Rock) Columba livia X X X X X X X X X X X X Common Wood Pigeon Columba palumbus X X X X X X X X X X X Eurasian Collared Dove Streptopelia decaocto X X X Common Swift Apus apus X X X X X X X X X X X X Barn Swallow Hirundo rustica X X X X X X X X X X X Common House Martin Delichon urbicum X X X X X X X X White Wagtail Motacilla alba X X -

Birdwatching Bingo Identification Sheet Hey Everyone! I Hope You Have Fun Playing Birdwatching Bingo with Your Family
Birdwatching Bingo Identification Sheet Hey Everyone! I hope you have fun playing Birdwatching Bingo with your family. You can even share it with friends and do it over social media. To help you on your adventure, here is an identification sheet with all the different birds listed on your bingo card. With the help of this sheet and some binoculars, you will be on your way to becoming a fantastic birdwatcher. Most of these birds you can see visiting a birdfeeder, but some you might have to go on a walk and look for with a parent. To play, download and print the bingo cards for each player. Using pennies, pieces of paper, or even a pencil, mark the card when you see and identify the different birds. Don’t be frustrated if you don’t finish the game in one sitting, this game might be completed over a couple days. A great tool to help with IDing birds is the app MERLIN. This is a free bird ID app for your phone from the Cornell Laboratory of Ornithology. The website, www.allaboutbirds.org, is a great source too. Blue Jay - Scientific Name: Cyanocitta cristata - Identifiable Markings: o Blue above, light grey below. Black and white markings on wings and tail. Larger than a robin, smaller than a crow. Crest and long tail. - Photo: Females/Males Look Similar Brown-headed Cowbird - Scientific Name: Molothrus ater - Identifiable Markings: o Stout bill. Short tail and stocky body. Males are glossy black with chocolate brown head. Females are grey-brown overall, without bold streaks, but slightly paler throat. -

Are European Starlings Breeding in the Azores Archipelago Genetically Distinct from Birds Breeding in Mainland Europe? Verónica C
Are European starlings breeding in the Azores archipelago genetically distinct from birds breeding in mainland Europe? Verónica C. Neves, Kate Griffiths, Fiona R. Savory, Robert W. Furness, Barbara K. Mable To cite this version: Verónica C. Neves, Kate Griffiths, Fiona R. Savory, Robert W. Furness, Barbara K. Mable. Are European starlings breeding in the Azores archipelago genetically distinct from birds breeding in mainland Europe?. European Journal of Wildlife Research, Springer Verlag, 2009, 56 (1), pp.95-100. 10.1007/s10344-009-0316-x. hal-00535248 HAL Id: hal-00535248 https://hal.archives-ouvertes.fr/hal-00535248 Submitted on 11 Nov 2010 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Eur J Wildl Res (2010) 56:95–100 DOI 10.1007/s10344-009-0316-x SHORT COMMUNICATION Are European starlings breeding in the Azores archipelago genetically distinct from birds breeding in mainland Europe? Verónica C. Neves & Kate Griffiths & Fiona R. Savory & Robert W. Furness & Barbara K. Mable Received: 6 May 2009 /Revised: 5 August 2009 /Accepted: 11 August 2009 /Published online: 29 August 2009 # Springer-Verlag 2009 Abstract The European starling (Sturnus vulgaris) has Keywords Azores . -

Bird Number Dynamics During the Post-Breeding Period at the Tömörd Bird Ringing Station, Western Hungary
THE RING 39 (2017) 10.1515/ring-2017-0002 BIRD NUMBER DYNAMICS DURING THE POST-BREEDING PERIOD AT THE TÖMÖRD BIRD RINGING STATION, WESTERN HUNGARY József Gyurácz1*, Péter Bánhidi2, József Góczán2, Péter Illés2, Sándor Kalmár2, Péter Koszorús2, Zoltán Lukács1, Csaba Németh2, László Varga2 ABSTRACT Gyurácz J., Bánhidi P., Góczán J., Illés P., Kalmár S., Koszorús P., Lukács Z., Németh C. and Varga L. 2017. Bird number dynamics during the post-breeding period at the Tömörd Bird Ringing Station, western Hungary. Ring 39: 23-82. The fieldwork, i.e. catching and ringing birds using mist-nets, was conducted at Tömörd Bird Ringing Station in western Hungary during the post-breeding migration seasons in 1998-2016. Altogether, 106,480 individuals of 133 species were ringed at the station. The aim of this paper was to publish basic information on passerine migration at this site. Migration phenology was described through annual and daily capture frequencies. Further- more, we provide the median date of the passage, the date of the earliest or latest capture, the peak migration season within the study period, and the countries where the birds monitored at the site were ringed or recovered abroad. To compare the catching dynamics for the fifty species with total captures greater than 200, a reference period was defined: from 5 Aug. to 5 Nov. 2001-2016. Some non-passerines that are more easily caught with mist-nets or that are caught occasionally were listed as well. The two superdominant spe- cies, the European Robin and the Eurasian Blackcap, with 14,377 and 13,926 total cap- tures, made up 27% of all ringed individuals. -

GHANA MEGA Rockfowl & Upper Guinea Specials Th St 29 November to 21 December 2011 (23 Days)
GHANA MEGA Rockfowl & Upper Guinea Specials th st 29 November to 21 December 2011 (23 days) White-necked Rockfowl by Adam Riley Trip Report compiled by Tour Leader David Hoddinott RBT Ghana Mega Trip Report December 2011 2 Trip Summary Our record breaking trip total of 505 species in 23 days reflects the immense birding potential of this fabulous African nation. Whilst the focus of the tour was certainly the rich assemblage of Upper Guinea specialties, we did not neglect the interesting diversity of mammals. Participants were treated to an astonishing 9 Upper Guinea endemics and an array of near-endemics and rare, elusive, localized and stunning species. These included the secretive and rarely seen White-breasted Guineafowl, Ahanta Francolin, Hartlaub’s Duck, Black Stork, mantling Black Heron, Dwarf Bittern, Bat Hawk, Beaudouin’s Snake Eagle, Congo Serpent Eagle, the scarce Long-tailed Hawk, splendid Fox Kestrel, African Finfoot, Nkulengu Rail, African Crake, Forbes’s Plover, a vagrant American Golden Plover, the mesmerising Egyptian Plover, vagrant Buff-breasted Sandpiper, Four-banded Sandgrouse, Black-collared Lovebird, Great Blue Turaco, Black-throated Coucal, accipiter like Thick- billed and splendid Yellow-throated Cuckoos, Olive and Dusky Long-tailed Cuckoos (amongst 16 cuckoo species!), Fraser’s and Akun Eagle-Owls, Rufous Fishing Owl, Red-chested Owlet, Black- shouldered, Plain and Standard-winged Nightjars, Black Spinetail, Bates’s Swift, Narina Trogon, Blue-bellied Roller, Chocolate-backed and White-bellied Kingfishers, Blue-moustached, -

Niche Analysis and Conservation of Bird Species Using Urban Core Areas
sustainability Article Niche Analysis and Conservation of Bird Species Using Urban Core Areas Vasilios Liordos 1,* , Jukka Jokimäki 2 , Marja-Liisa Kaisanlahti-Jokimäki 2, Evangelos Valsamidis 1 and Vasileios J. Kontsiotis 1 1 Department of Forest and Natural Environment Sciences, International Hellenic University, 66100 Drama, Greece; [email protected] (E.V.); [email protected] (V.J.K.) 2 Arctic Centre, University of Lapland, 96101 Rovaniemi, Finland; jukka.jokimaki@ulapland.fi (J.J.); marja-liisa.kaisanlahti@ulapland.fi (M.-L.K.-J.) * Correspondence: [email protected] Abstract: Knowing the ecological requirements of bird species is essential for their successful con- servation. We studied the niche characteristics of birds in managed small-sized green spaces in the urban core areas of southern (Kavala, Greece) and northern Europe (Rovaniemi, Finland), during the breeding season, based on a set of 16 environmental variables and using Outlying Mean Index, a multivariate ordination technique. Overall, 26 bird species in Kavala and 15 in Rovaniemi were recorded in more than 5% of the green spaces and were used in detailed analyses. In both areas, bird species occupied different niches of varying marginality and breadth, indicating varying responses to urban environmental conditions. Birds showed high specialization in niche position, with 12 species in Kavala (46.2%) and six species in Rovaniemi (40.0%) having marginal niches. Niche breadth was narrower in Rovaniemi than in Kavala. Species in both communities were more strongly associated either with large green spaces located further away from the city center and having a high vegetation cover (urban adapters; e.g., Common Chaffinch (Fringilla coelebs), European Greenfinch (Chloris Citation: Liordos, V.; Jokimäki, J.; chloris Cyanistes caeruleus Kaisanlahti-Jokimäki, M.-L.; ), Eurasian Blue Tit ( )) or with green spaces located closer to the city center Valsamidis, E.; Kontsiotis, V.J. -

Federal Register/Vol. 85, No. 74/Thursday, April 16, 2020/Notices
21262 Federal Register / Vol. 85, No. 74 / Thursday, April 16, 2020 / Notices acquisition were not included in the 5275 Leesburg Pike, Falls Church, VA Comment (1): We received one calculation for TDC, the TDC limit would not 22041–3803; (703) 358–2376. comment from the Western Energy have exceeded amongst other items. SUPPLEMENTARY INFORMATION: Alliance, which requested that we Contact: Robert E. Mulderig, Deputy include European starling (Sturnus Assistant Secretary, Office of Public Housing What is the purpose of this notice? vulgaris) and house sparrow (Passer Investments, Office of Public and Indian Housing, Department of Housing and Urban The purpose of this notice is to domesticus) on the list of bird species Development, 451 Seventh Street SW, Room provide the public an updated list of not protected by the MBTA. 4130, Washington, DC 20410, telephone (202) ‘‘all nonnative, human-introduced bird Response: The draft list of nonnative, 402–4780. species to which the Migratory Bird human-introduced species was [FR Doc. 2020–08052 Filed 4–15–20; 8:45 am]‘ Treaty Act (16 U.S.C. 703 et seq.) does restricted to species belonging to biological families of migratory birds BILLING CODE 4210–67–P not apply,’’ as described in the MBTRA of 2004 (Division E, Title I, Sec. 143 of covered under any of the migratory bird the Consolidated Appropriations Act, treaties with Great Britain (for Canada), Mexico, Russia, or Japan. We excluded DEPARTMENT OF THE INTERIOR 2005; Pub. L. 108–447). The MBTRA states that ‘‘[a]s necessary, the Secretary species not occurring in biological Fish and Wildlife Service may update and publish the list of families included in the treaties from species exempted from protection of the the draft list. -

Science for Saving Species Research Findings Factsheet Project 4.2
Science for Saving Species Research findings factsheet Project 4.2 Assessing the impacts of invasive species: Hollow-nesting birds in Tasmania In brief Background Predicting the impacts of invasive Invasive alien birds are found explore the known and theoretical species is difficult at large spatial across many areas of Australia. interaction network between the scales. This is because the interactions Many of these introduced birds cavity breeding birds in Tasmania, between invasive species and native use cavities, an important breeding and to identify which native species species vary across different species, resource for cavity nesting species. are likely impacted by the addition between different locations, over In Tasmania alone there are 27 of non-native species. time and in relation to other pressures species of hollow-nesting birds, We discovered that, overall, such as habitat loss, extensive fires, including three threatened species native hollow-nesting species climatic events and drought. and seven invasive hollow-nesting are likely facing increased levels bird species. The logging of big old Given that conservation and of competition for nesting sites trees with cavities and the addition management work is almost always as a result of non-native species of invasive species has likely led conducted under limited budgets and introductions. Such competition to increased competition over time, being able to quantify where is likely to decrease breeding the limited resource. However, and when invasive species are having opportunities for native species, the impact of most of these a significant impact on local species including some of Tasmania’s invasive species has not been is vital for effectively managing and threatened and endemic species.