Unravelling the Subfields of the Hippocampal Head Using 7-Tesla Structural MRI
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Reversal Sign Daniel Gaete,1 Antonio Lopez-Rueda2
Images in… BMJ Case Reports: first published as 10.1136/bcr-2014-204419 on 16 May 2014. Downloaded from The reversal sign Daniel Gaete,1 Antonio Lopez-Rueda2 1Clinica Alemana de Santiago, DESCRIPTION Santiago, Chile A 75-year-old man with a history of chronic 2Hospital Clinic i Provincial de Barcelona, Barcelona, Spain obstructive pulmonary disease was found in cardio- pulmonary arrest. After successful resuscitation the Correspondence to patient was transferred to our institution. On Antonio Lopez-Rueda, arrival, a non-enhanced brain CT was performed to [email protected] assess brain damage, which showed signs of diffuse Accepted 18 April 2014 cerebral oedema, with effacement of the cerebral sulci, sulcal hyperdensity and decreased attenuation of deep and cortical grey matter which appears hypodense in comparison to the white matter, a finding referred to as the ‘reversal sign’ (figures 1 and 2). These injuries were secondary to global brain ischaemia. In less than 8 h, the patient devel- oped multiple organ dysfunction syndrome and was pronounced dead. Cardiopulmonary arrest may lead to diffuse hypoxic ischaemic brain injury. Initially, unen- hanced brain CT may show subtle hypodensity of Figure 2 Coronal reformatted image of the same the basal ganglia and insular cortex, with efface- patient showing the reversal sign, which is also present ment of the basal cisterns. When diffuse brain in the cerebellum. oedema develops, the findings become more obvious, with effacement of the sulci and cisterns, The reversal sign reflects a diffuse hypoxic and loss of the grey matter–white matter differenti- ischaemic cerebral injury, with irreversible brain ation. -

Histological Underpinnings of Grey Matter Changes in Fibromyalgia Investigated Using Multimodal Brain Imaging
1090 • The Journal of Neuroscience, February 1, 2017 • 37(5):1090–1101 Neurobiology of Disease Histological Underpinnings of Grey Matter Changes in Fibromyalgia Investigated Using Multimodal Brain Imaging X Florence B. Pomares,1,2 Thomas Funck,3 Natasha A. Feier,1 XSteven Roy,1 X Alexandre Daigle-Martel,4 XMarta Ceko,5 Sridar Narayanan,3 XDavid Araujo,3 Alexander Thiel,6,7 Nikola Stikov,4,8 Mary-Ann Fitzcharles,9,10 and Petra Schweinhardt1,2,6,11 1Alan Edwards Centre for Research on Pain, McGill University, Montreal, Quebec H3A 0C7, Canada, 2Faculty of Dentistry, McGill University, Montreal, Quebec H3A 0C7, Canada, 3McConnell Brain Imaging Centre, Montreal Neurological Institute, McGill University, Montreal, Quebec, H3A 2B4, Canada, 4Institute for Biomedical Engineering, E´cole Polytechnique, Montreal, Quebec H3T 1J4, Canada, 5Institute of Cognitive Science, University of Colorado, Boulder, Colorado 80309, 6Department of Neurology & Neurosurgery, McGill University, Montreal, Quebec H3A 0C7, Canada, 7Jewish General Hospital, Montreal, Quebec H3T 1E2, Canada, 8Montreal Heart Institute, Montreal, Quebec, H1T 1C8, Canada, 9Division of Rheumatology, McGill University Health Centre, Montreal, Quebec H3G 1A4, Canada, 10Alan Edwards Pain Management Unit, McGill University Health Centre, Montreal, Quebec H3G 1A4, Canada, and 11Interdisciplinary Spinal Research, Department of Chiropractic Medicine, University Hospital Balgrist, 8008 Zurich, Switzerland Chronic pain patients present with cortical gray matter alterations, observed with anatomical magnetic resonance (MR) imaging. Reduced regional gray matter volumes are often interpreted to reflect neurodegeneration, but studies investigating the cellular origin of gray matter changesarelacking.Weusedmultimodalimagingtocompare26postmenopausalwomenwithfibromyalgiawith25healthycontrols(agerange: 50–75 years) to test whether regional gray matter volume decreases in chronic pain are associated with compromised neuronal integrity. -

OBSERVATIONS on PARTIAL REMOVAL of the POST-CENTRAL GYRUS for PAIN by WALPOLE LEWIN and C
J Neurol Neurosurg Psychiatry: first published as 10.1136/jnnp.15.3.143 on 1 August 1952. Downloaded from J. Neurol. Neurosurg. Psychiat., 1952, 15, 143. OBSERVATIONS ON PARTIAL REMOVAL OF THE POST-CENTRAL GYRUS FOR PAIN BY WALPOLE LEWIN and C. G. PHILLIPS From the Nuffield Department ofSurgery and the University Laboratory ofPhysiology, Oxford The role of the cerebral cortex in the conscious one case where stimulation of the sensory cortex appreciation of pain has interested neurologists for produced pain in the phantom leg. many years. Head and Holmes (1911) considered In this paper we wish to record three cases in that pain entered consciousness at thalamic level, which partial resection of the post-central gyrus and more recently Penfleld (1947) stated that was undertaken for the relief of pain. In the first 6. .no removal of cortex anywhere can prevent patient the pain developed during an unusual, pain from being felt and only very rarely does a prolonged, sensory painful aura in traumatic patient use the word pain to describe the result of epilepsy, the second patient had intractable pain in cortical stimulation ", and, he goes on, " it is a phantom foot, and the third had a painful thigh obvious therefore that the pathway of pain conduc- stump. In all three, electrical stimulation of the Protected by copyright. tion reaches the thalamus and consciousness appropriate area of the post-central gyrus repro- without essential conduction to the cortex ". duced the pain complained of by the patient and Adrian (1941) found that no impulses reached the relief followed the removal of this area of cortex. -

Translingual Neural Stimulation with the Portable Neuromodulation
Translingual Neural Stimulation With the Portable Neuromodulation Stimulator (PoNS®) Induces Structural Changes Leading to Functional Recovery In Patients With Mild-To-Moderate Traumatic Brain Injury Authors: Jiancheng Hou,1 Arman Kulkarni,2 Neelima Tellapragada,1 Veena Nair,1 Yuri Danilov,3 Kurt Kaczmarek,3 Beth Meyerand,2 Mitchell Tyler,2,3 *Vivek Prabhakaran1 1. Department of Radiology, School of Medicine and Public Health, University of Wisconsin-Madison, Madison, Wisconsin, USA 2. Department of Biomedical Engineering, University of Wisconsin-Madison, Madison, Wisconsin, USA 3. Department of Kinesiology, University of Wisconsin-Madison, Madison, Wisconsin, USA *Correspondence to [email protected] Disclosure: Dr Tyler, Dr Danilov, and Dr Kaczmarek are co-founders of Advanced Neurorehabilitation, LLC, which holds the intellectual property rights to the PoNS® technology. Dr Tyler is a board member of NeuroHabilitation Corporation, a wholly- owned subsidiary of Helius Medical Technologies, and owns stock in the corporation. The other authors have declared no conflicts of interest. Acknowledgements: Professional medical writing and editorial assistance were provided by Kelly M. Fahrbach, Ashfield Healthcare Communications, part of UDG Healthcare plc, funded by Helius Medical Technologies. Dr Tyler, Dr Kaczmarek, Dr Danilov, Dr Hou, and Dr Prabhakaran were being supported by NHC-TBI-PoNS-RT001. Dr Hou, Dr Kulkarni, Dr Nair, Dr Tellapragada, and Dr Prabhakaran were being supported by R01AI138647. Dr Hou and Dr Prabhakaran were being supported by P01AI132132, R01NS105646. Dr Kulkarni was being supported by the Clinical & Translational Science Award programme of the National Center for Research Resources, NCATS grant 1UL1RR025011. Dr Meyerand, Dr Prabhakaran, Dr Nair was being supported by U01NS093650. -

Neocortex: Consciousness Cerebellum
Grey matter (chips) White matter (the wiring: the brain mainly talks to itself) Neocortex: consciousness Cerebellum: unconscious control of posture & movement brains 1. Golgi-stained section of cerebral cortex 2. One of Ramon y Cajal’s faithful drawings showing nerve cell diversity in the brain cajal Neuropil: perhaps 1 km2 of plasma membrane - a molecular reaction substrate for 1024 voltage- and ligand-gated ion channels. light to Glia: 3 further cell types 1. Astrocytes: trophic interface with blood, maintain blood brain barrier, buffer excitotoxic neurotransmitters, support synapses astros Oligodendrocytes: myelin insulation oligos Production persists into adulthood: radiation myelopathy 3. Microglia: resident macrophages of the CNS. Similarities and differences with Langerhans cells, the professional antigen-presenting cells of the skin. 3% of all cells, normally renewed very slowly by division and immigration. Normal Neurosyphilis microglia Most adult neurons are already produced by birth Peak synaptic density by 3 months EMBRYONIC POSTNATAL week: 0 6 12 18 24 30 36 Month: 0 6 12 18 24 30 36 Year: 4 8 12 16 20 24 Cell birth Migration 2* Neurite outgrowth Synaptogenesis Myelination 1* Synapse elimination Modified from various sources inc: Andersen SL Neurosci & Biobehav Rev 2003 Rakic P Nat Rev Neurosci 2002 Bourgeois Acta Pediatr Suppl 422 1997 timeline 1 Synaptogenesis 100% * Rat RTH D BI E A Density of synapses in T PUBERTY primary visual cortex H at different times post- 0% conception. 100% (logarithmic scale) RTH Cat BI D E A T PUBERTY H The density values equivalent 0% to 100% vary between species 100% but in Man the peak value is Macaque 6 3 RTH 350 x10 synapses per mm BI D E PUBERTY A T The peak rate of synapse H formation is at birth in the 0% macaque: extrapolating to 100% the entire cortex, this Man RTH BI amounts to around 800,000 D E synapses formed per sec. -
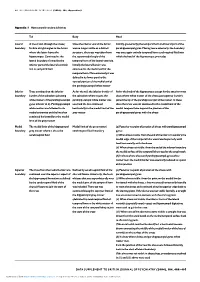
Appendix 1..1
BRITISH JOURNAL OF PSYCHIATRY (2004), 184, Appendix/1 Appendix 1 Hippocampal boundary definitions TailBody Head LateralLateral A line drawn through the alveus/ Once the inferior end of the fornix Initially (posteriorly) the temporal horn and lateral part of the boundary fimbria at right angles to the fornix was no longer visible as a distinct parahippocampal gyrus. Moving more anteriorly, the boundary where the latter leaves the structure, the trace was taken from was once again entirely temporal horn cerebrospinal fluid into hippocampus. Continued as the the superomedial angle of the which the head of the hippocampus protrudes lateral boundary, formed by the temporal horn of the lateral ventricle. inferior part of the lateral ventricle Initially the lateral border was and its temporal horn continued as the medial wall of the temporal horn. More anteriorly it was defined in its lower part by the vertical portion of the medial end of thetheparahippocampalwhitematter parahippocampal white matter Inferior Trace continued on the inferior As for the tail, the inferior border of As for the body of the hippocampus except for the anterior-most boundary border of the subiculum (abutting the subiculum where it joins the slices where white matter of the alveus (see superior border) white matter of the parahippocampal parahippocampal white matter was joined the tip of the parahippocampal white matter. In these gyrus inferior to it). Parahippocampal used and the line continued slices the trace was not continued to the medial limit of the white matter -

The Anatomy of First-Episode and Chronic Schizophrenia: an Anatomical Likelihood Estimation Meta-Analysis
Article The Anatomy of First-Episode and Chronic Schizophrenia: An Anatomical Likelihood Estimation Meta-Analysis Ian Ellison-Wright, M.R.C.P. Objective: The authors sought to map schizophrenia and chronic schizophrenia, gray matter changes in first-episode including gray matter decreases in the David C. Glahn, Ph.D. schizophrenia and to compare these with thalamus, the left uncus/amygdala re- the changes in chronic schizophrenia. gion, the insula bilaterally, and the ante- They postulated that the data would rior cingulate. In the comparison of first- Angela R. Laird, Ph.D. show a progression of changes from hip- episode schizophrenia and chronic pocampal deficits in first-episode schizo- schizophrenia, decreases in gray matter Sarah M. Thelen, B.S. phrenia to include volume reductions in volume were detected in first-episode the amygdala and cortical gray matter in schizophrenia but not in chronic schizo- Ed Bullmore, Ph.D. chronic schizophrenia. phrenia in the caudate head bilaterally; decreases were more widespread in corti- Method: A systematic search was con- cal regions in chronic schizophrenia. ducted for voxel-based structural MRI studies of patients with first-episode Conclusions: Anatomical changes in schizophrenia and chronic schizophrenia first-episode schizophrenia broadly coin- in relation to comparison groups. Meta- cide with a basal ganglia-thalamocortical analyses of the coordinates of gray matter circuit. These changes include bilateral re- differences were carried out using ana- ductions in caudate head gray matter, tomical likelihood estimation. Maps of which are absent in chronic schizophre- gray matter changes were constructed, nia. Comparing first-episode schizophre- and subtraction meta-analysis was used nia and chronic schizophrenia, the au- to compare them. -

Age Differentiation Within Grey Matter, White Matter and Between Memory and White Matter in an Adult Lifespan Cohort
bioRxiv preprint doi: https://doi.org/10.1101/148452; this version posted February 26, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. Age differentiation within grey matter, white matter and between memory and white matter in an adult lifespan cohort Susanne M.M. de Mooij*,1, Richard N.A. Henson2, Lourens J. Waldorp1, Cam-CAN3, & Rogier A. Kievit2 *corresponding author: [email protected], 1Department of Psychology, University of Amsterdam, Roetersstraat 15, 1018 WB Amsterdam, The Netherlands, 2MRC Cognition and Brain Sciences Unit, 15 Chaucer Road, Cambridge CB2 7EF, UK, 3Cambridge Centre for Ageing and Neuroscience (Cam-CAN), University of Cambridge and MRC Cognition and Brain Sciences Unit, Cambridge, UK Abstract It is well-established that brain structures and cognitive functions change across the lifespan. A longstanding hypothesis called age differentiation additionally posits that the relations between cognitive functions also change with age. To date however, evidence for age-related differentiation is mixed, and no study has examined differentiation of the relationship between brain and cognition. Here we use multi-group Structural Equation Modeling and SEM Trees to study differences within and between brain and cognition across the adult lifespan (18-88 years) in a large (N>646, closely matched across sexes), population-derived sample of healthy human adults from the Cambridge Centre for Ageing and Neuroscience (www.cam- can.org). After factor analyses of grey-matter volume (from T1- and T2-weighted MRI) and white-matter organisation (fractional anisotropy from Diffusion-weighted MRI), we found evidence for differentiation of grey and white matter, such that the covariance between brain factors decreased with age. -

Brain Anatomy
BRAIN ANATOMY Adapted from Human Anatomy & Physiology by Marieb and Hoehn (9th ed.) The anatomy of the brain is often discussed in terms of either the embryonic scheme or the medical scheme. The embryonic scheme focuses on developmental pathways and names regions based on embryonic origins. The medical scheme focuses on the layout of the adult brain and names regions based on location and functionality. For this laboratory, we will consider the brain in terms of the medical scheme (Figure 1): Figure 1: General anatomy of the human brain Marieb & Hoehn (Human Anatomy and Physiology, 9th ed.) – Figure 12.2 CEREBRUM: Divided into two hemispheres, the cerebrum is the largest region of the human brain – the two hemispheres together account for ~ 85% of total brain mass. The cerebrum forms the superior part of the brain, covering and obscuring the diencephalon and brain stem similar to the way a mushroom cap covers the top of its stalk. Elevated ridges of tissue, called gyri (singular: gyrus), separated by shallow groves called sulci (singular: sulcus) mark nearly the entire surface of the cerebral hemispheres. Deeper groves, called fissures, separate large regions of the brain. Much of the cerebrum is involved in the processing of somatic sensory and motor information as well as all conscious thoughts and intellectual functions. The outer cortex of the cerebrum is composed of gray matter – billions of neuron cell bodies and unmyelinated axons arranged in six discrete layers. Although only 2 – 4 mm thick, this region accounts for ~ 40% of total brain mass. The inner region is composed of white matter – tracts of myelinated axons. -

Nervous System Pt 3
Write this down… Homework 2 Study Guide (Synapses) Due at the beginning of lab this week Front and back TASS M&W 1-2pm Willamette Hall 204 Thought Question… When you have one of your mandibular teeth worked on at the dentist and he gives you a shot to deaden half of your mouth, what division of the nervous system is being affected by the lidocaine? What do you think it’s mode of action is? Hint: Remember Physio-EX in lab? Is it affecting a cranial or spinal nerve? The Nervous System THE CENTRAL NERVOUS SYSTEM, T H E B R A I N Introduction Integration Memory Learning Sensation and perception Neural Tissue - Definitions White matter versus Gray matter Fiber Bundles Nerves versus Tracts Nerve Cell Bodies Nucleus versus Ganglion White and Gray Matter Central cavity surrounded by a gray matter core External white matter composed of myelinated fiber tracts Brain has additional areas of gray matter not present in spinal cord Central cavity Cortex of gray matter Migratory Inner gray pattern of matter neurons Cerebrum Outer white Cerebellum matter Gray matter Region of cerebellum Central cavity Inner gray matter Outer white matter Gray matter Brain stem Central cavity Outer white matter Inner gray matter Spinal cord Copyright © 2010 Pearson Education, Inc. Figure 12.4 Brain Similar pattern with additional areas of gray matter The Brain Conscious perception Internal regulation Average adult male 3.5 lbs Same brain mass Average adult female 3.2 lbs to body mass ratio! Brain Regions 4 Adult brain regions 1. Cerebral hemispheres (cerebrum) 2. -

Grey Matter Matters: Teaching Strategies for the Brain Compatible
Teaching & Professional Practice Grey matter matters Teaching strategies for the brain compatible classroom Andrea Thompson Primary teacher, Avondale Schools, Cooranbong, NSW The brain is extremely complex. The brain has discovered in our universe” (MacDonald, 2008, the amazing ability to reshape and reorganise p. 18). When we remember that “God’s intelligence its neural networks, depending on increased or is the basis of human intelligence” (Sire, 1977, p. 35), decreased use, making it malleable or ‘plastic’. the complexity of the brain is not surprising. This plasticity allows for incredible changes to take place, which were once thought impossible. The brain’s ability to rewire This article explores current research in this Our brain has been designed by a loving God, with area and offers brain compatible strategies the ability to change its own wiring to make it more that teachers can employ in the classroom to efficient. Neuroplasticity is the word that describes make learning more efficient, to raise student the brain’s ability to rewire and can be defined as achievement, and to facilitate a healthy learning the “genetically driven overproduction of synapses environment. and the environmentally driven maintenance and pruning of synaptic connections” (Cicchetti and Christians have long agreed with the Psalmist Curtis, 2006, as cited by Willis and Kappan, 2008, that humans are “so wonderfully complex” (Psalm p. 4). The brain’s plasticity allows it to reshape and Our brain is 139:4 NLT), yet it is only comparatively recently reorganise networks, depending on increased or constantly that advances in neuroscience have allowed decreased use. This makes the brain malleable and learning how researchers to observe how complex the human able to change. -

The Heart Side of Brain Neuromodulation
The heart side of brain neuromodulation rsta.royalsocietypublishing.org Simone Rossi1, Emiliano Santarnecchi1,2, Gaetano Valenza3,4,5 and Monica Ulivelli1 Review 1Department of Medicine, Surgery and Neuroscience, Unit of Neurology and Clinical Neurophysiology, Brain Investigation and Cite this article: Rossi S, Santarnecchi E, Neuromodulation Lab. (Si-BIN Lab.), Azienda Ospedaliera Valenza G, Ulivelli M. 2016 The heart side of Universitaria Senese, University of Siena, 53100 Siena, Italy Phil.Trans.R.Soc.A brain neuromodulation. 2Berenson–Allen Center for Noninvasive Brain Stimulation, 374:20150187. Department of Neurology, Beth Israel Deaconess Medical Center, http://dx.doi.org/10.1098/rsta.2015.0187 Boston, MA 02215, USA 3Department of Information Engineering, and Research Center Accepted: 11 January 2016 E. Piaggio, University of Pisa, 56122 Pisa, Italy 4Neuroscience Statistics Research Lab, Harvard Medical School, One contribution of 16 to a theme issue Massachusetts General Hospital, Boston, MA 02115, USA ‘Uncovering brain–heart information through 5Massachusetts Institute of Technology, Cambridge, MA 02139, USA advanced signal and image processing’. SR, 0000-0001-6697-9459 Subject Areas: biomedical engineering Neuromodulation refers to invasive, minimally invasive or non-invasive techniques to stimulate Keywords: discrete cortical or subcortical brain regions with deep brain stimulation, vagal nerve therapeutic purposes in otherwise intractable patients: for example, thousands of advanced Parkinsonian stimulation, repetitive transcranial