Improved Sampling at the Subspecies Level Solves a Taxonomic Dilemma Â
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Sichuan, China
Tropical Birding: Sichuan (China). Custom Tour Report A Tropical Birding custom tour SICHUAN, CHINA : (Including the Southern Shans Pre-tour Extension) WHITE-THROATED TIT One of 5 endemic tits recorded on the tour. 21 May – 12 June, 2010 Tour Leader: Sam Woods All photos were taken by Sam Woods/Tropical Birding on this tour, except one photo. www.tropicalbirding.com [email protected] 1-409-515-0514 Tropical Birding: Sichuan (China). Custom Trip Report The Central Chinese province of Sichuan provided some notable challenges this year: still recovering from the catastrophic “Wenchuan 5.12” earthquake of 2008, the area is undergoing massive reconstruction. All very positive for the future of this scenically extraordinary Chinese region, but often a headache for tour arrangements, due to last minute traffic controls leading us to regularly rethink our itinerary in the Wolong area in particular, that was not far from the epicenter of that massive quake. Even in areas seemingly unaffected by the quake, huge road construction projects created similar challenges to achieving our original planned itinerary. However, in spite of regular shuffling and rethinking, the itinerary went ahead pretty much as planned with ALL sites visited. Other challenges came this year in the form of heavy regular rains that plagued us at Wawu Shan and low cloud that limited visibility during our time around the breathtaking Balang Mountain in the Wolong region. With some careful trickery, sneaking our way through week-long road blocks under cover of darkness, birding through thick and thin (mist, cloud and rains) we fought against all such challenges and came out on top. -

Wild China – Sichuan's Birds & Mammals
Wild China – Sichuan’s Birds & Mammals Naturetrek Tour Report 9 - 24 November 2019 Golden snub-nosed Monkey White-browed Rosefinch Himalayan Vulture Red Panda Report and images compiled by Barrie Cooper Naturetrek Mingledown Barn Wolf's Lane Chawton Alton Hampshire GU34 3HJ UK T: +44 (0)1962 733051 E: [email protected] W: www.naturetrek.co.uk Tour Report Wild China – Sichuan’s Birds & Mammals Tour participants: Barrie Cooper (leader), Sid Francis (Local Guide), with six Naturetrek clients. Summary Sichuan is a marvellous part of China with spectacular scenery, fine food and wonderful wildlife. We had a rich variety of mammals and birds on this trip. Mammals included Red Panda, Golden Snub-nosed Monkey, the endemic Chinese Desert Cat, Pallas’s Cat on two days, including during the daytime. The “golden fleece animal” –Takin never fails to attract the regular attention of cameras. A good range of bird species, including several endemics, was also recorded and one lucky group member now has Temminck’s Tragopan on his list. Day 1 Saturday 9th November The Cathay Pacific flight from Heathrow to Hong Kong had its usual good selection of films and decent food. Day 2 Sunday 10th November The group of three from London, became seven when we met the others at the gate for the connecting flight to Chengdu the following morning. Unfortunately, the flight was delayed by an hour but we had the compensation of warm, sunny weather at Chengdu where we met up with our local guide Sid. After a couple of hours of driving, we arrived at our hotel in Dujiangyan. -
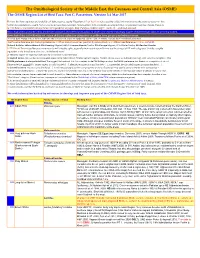
OSME List V3.4 Passerines-2
The Ornithological Society of the Middle East, the Caucasus and Central Asia (OSME) The OSME Region List of Bird Taxa: Part C, Passerines. Version 3.4 Mar 2017 For taxa that have unproven and probably unlikely presence, see the Hypothetical List. Red font indicates either added information since the previous version or that further documentation is sought. Not all synonyms have been examined. Serial numbers (SN) are merely an administrative conveninence and may change. Please do not cite them as row numbers in any formal correspondence or papers. Key: Compass cardinals (eg N = north, SE = southeast) are used. Rows shaded thus and with yellow text denote summaries of problem taxon groups in which some closely-related taxa may be of indeterminate status or are being studied. Rows shaded thus and with white text contain additional explanatory information on problem taxon groups as and when necessary. A broad dark orange line, as below, indicates the last taxon in a new or suggested species split, or where sspp are best considered separately. The Passerine Reference List (including References for Hypothetical passerines [see Part E] and explanations of Abbreviated References) follows at Part D. Notes↓ & Status abbreviations→ BM=Breeding Migrant, SB/SV=Summer Breeder/Visitor, PM=Passage Migrant, WV=Winter Visitor, RB=Resident Breeder 1. PT=Parent Taxon (used because many records will antedate splits, especially from recent research) – we use the concept of PT with a degree of latitude, roughly equivalent to the formal term sensu lato , ‘in the broad sense’. 2. The term 'report' or ‘reported’ indicates the occurrence is unconfirmed. -

EUROPEAN BIRDS of CONSERVATION CONCERN Populations, Trends and National Responsibilities
EUROPEAN BIRDS OF CONSERVATION CONCERN Populations, trends and national responsibilities COMPILED BY ANNA STANEVA AND IAN BURFIELD WITH SPONSORSHIP FROM CONTENTS Introduction 4 86 ITALY References 9 89 KOSOVO ALBANIA 10 92 LATVIA ANDORRA 14 95 LIECHTENSTEIN ARMENIA 16 97 LITHUANIA AUSTRIA 19 100 LUXEMBOURG AZERBAIJAN 22 102 MACEDONIA BELARUS 26 105 MALTA BELGIUM 29 107 MOLDOVA BOSNIA AND HERZEGOVINA 32 110 MONTENEGRO BULGARIA 35 113 NETHERLANDS CROATIA 39 116 NORWAY CYPRUS 42 119 POLAND CZECH REPUBLIC 45 122 PORTUGAL DENMARK 48 125 ROMANIA ESTONIA 51 128 RUSSIA BirdLife Europe and Central Asia is a partnership of 48 national conservation organisations and a leader in bird conservation. Our unique local to global FAROE ISLANDS DENMARK 54 132 SERBIA approach enables us to deliver high impact and long term conservation for the beneit of nature and people. BirdLife Europe and Central Asia is one of FINLAND 56 135 SLOVAKIA the six regional secretariats that compose BirdLife International. Based in Brus- sels, it supports the European and Central Asian Partnership and is present FRANCE 60 138 SLOVENIA in 47 countries including all EU Member States. With more than 4,100 staf in Europe, two million members and tens of thousands of skilled volunteers, GEORGIA 64 141 SPAIN BirdLife Europe and Central Asia, together with its national partners, owns or manages more than 6,000 nature sites totaling 320,000 hectares. GERMANY 67 145 SWEDEN GIBRALTAR UNITED KINGDOM 71 148 SWITZERLAND GREECE 72 151 TURKEY GREENLAND DENMARK 76 155 UKRAINE HUNGARY 78 159 UNITED KINGDOM ICELAND 81 162 European population sizes and trends STICHTING BIRDLIFE EUROPE GRATEFULLY ACKNOWLEDGES FINANCIAL SUPPORT FROM THE EUROPEAN COMMISSION. -

Population Genetics and Phylogeography of the Pygmy Nuthatch in Southern California
California State University, San Bernardino CSUSB ScholarWorks Theses Digitization Project John M. Pfau Library 2006 Population genetics and phylogeography of the pygmy nuthatch in Southern California Thomas Alan Benson Follow this and additional works at: https://scholarworks.lib.csusb.edu/etd-project Part of the Population Biology Commons Recommended Citation Benson, Thomas Alan, "Population genetics and phylogeography of the pygmy nuthatch in Southern California" (2006). Theses Digitization Project. 2973. https://scholarworks.lib.csusb.edu/etd-project/2973 This Thesis is brought to you for free and open access by the John M. Pfau Library at CSUSB ScholarWorks. It has been accepted for inclusion in Theses Digitization Project by an authorized administrator of CSUSB ScholarWorks. For more information, please contact [email protected]. POPULATION GENETICS AND PHYLOGEOGRAPHY OF THE PYGMY NUTHATCH IN SOUTHERN CALIFORNIA A Thesis Presented to the Faculty of California State University, San Bernardino In Partial Fulfillment of the Requirements for the Degree Master of Science in Biology by Thomas Alan Benson June 2006 POPULATION GENETICS AND PHYLOGEOGRAPHY OF THE PYGMY NUTHATCH IN SOUTHERN CALIFORNIA A Thesis Presented to the Faculty of California State University, San Bernardino by Thomas Alan Benson June 2006 Approved by: Anthony JHeitc^lf, PhpfC , Chair, Biology Date Richard Fehn,(Ph.D. James Ferrari, Ph.D. ABSTRACT I examined the population genetic structure, phylogeography, and subspecies structure of pygmy nuthatch (Sitta pygmaea') populations in southern California using mitochondrial DNA sequence data. Population genetic analyses indicate that pygmy nuthatch populations fragmented in the disjunct mountain ranges of southern California exhibit low but significant levels of genetic differentiation. -

Nature Holidays and Wildlife Photography Tours Welcome to Our New Brochure Showing a Selection of 2012 Tours
2012 Nature holidays and wildlife photography tours Welcome to our new brochure showing a selection of 2012 tours. Those presented here are about half of the tours we have to offer – our full tour program can be found at our website www.spatiawildlife.com After another successful year we are now ready for some major changes, for better, positive steps forward. First of all we have added more tours, to destinations other than our home country of Bulgaria, gradually expanding the areas we cover. There are two new birdwatching tours to Jordan and Ethiopia. While the first one is a pretty much underlooked country, the second one is a well established birdwatching destination offering an amazing variety of species, and not only for birds! Together with these two we are offering again dates for Turkey where we have run two successful trips last May as well as tour to Azerbaijan, which is definitely the least visited country in the Western Palearctic. Our first tour there last June revealed the country’s big potential and we are looking forward to returning next year. On the butterfly front beside the successful Butterfly of the Balkans tour where this year’s group saw an amazing 143 species, we have also prepared two new itineraries. Indeed, both by request of our clients. First one is focused on the very early spring butterflies of Bulgaria and Greece which are always missed on the later tours in June and July. The second tour is in the prime butterfly time, but is focused on some rare species which are not easy to seen on the classical tours. -

Type Specimens of Birds in the American Museum of Natural History
L Scientific Publications of the American Museum of Natural History e Croy American Museum Novitates TyPe SPeCIMeNS oF BIrDS IN THe Bulletin of the American Museum of Natural History : Anthropological Papers of the American Museum of Natural History T AMERICAN MUSeUM oF NATUrAL HISTORY y P Publications Committee e SP PArT 8. PASSERIFORMeS: Robert S. Voss, Chair e Board of Editors CIM PACHyCePHALIDAe, AeGITHALIDAe, reMIZIDAe, Jin Meng, Paleontology e Lorenzo Prendini, Invertebrate Zoology NS PArIDAe, SITTIDAe, NEOSITTIDAe, CERTHIIDAe, Robert S. Voss, Vertebrate Zoology o rHABDORNITHIDAe, CLIMACTERIDAe, DICAeIDAe, Peter M. Whiteley, Anthropology F BI PArDALoTIDAe, AND NeCTArINIIDAe Managing Editor r DS: DS: Mary Knight 8. PAS M Ary L eCroy Submission procedures can be found at http://research.amnh.org/scipubs S ER Complete lists of all issues of Novitates and Bulletin are available on the web (http:// IF digitallibrary.amnh.org/dspace). Inquire about ordering printed copies via e-mail from OR [email protected] or via standard mail from: M e American Museum of Natural History—Scientific Publications, S Central Park West at 79th St., New York, NY 10024. This paper meets the requirements of ANSI/NISO Z39.48-1992 (permanence of paper). AMNH BULL On the cover: The type specimen of Pachycephala nudigula Hartert, 1897, shown here in a lithograph by J.G. e TIN 333 Keulemans (Novitates Zoologicae, 1897, 4: pl. 3, fig.3), was collected by Alfred Everett on Flores Island, Indonesia, in October 1896. The bare, deep red throat, unique in the genus, occurs only in the adult male and is inflated when he sings. -

FIELD GUIDES BIRDING TOURS: China: Manchuria & the Tibetan Plateau 2013
Field Guides Tour Report China: Manchuria & the Tibetan Plateau 2013 May 4, 2013 to May 25, 2013 Dave Stejskal & Jesper Hornskov For our tour description, itinerary, past triplists, dates, fees, and more, please VISIT OUR TOUR PAGE. Of the five crane species we saw on the tour, the Red-crowned Crane is probably the rarest in China, though they still are fairly common in Japan. (Photo by guide Dave Stejskal) Although this was Field Guides' third tour to China with co-leader Jesper Hornskov as our host, this was the first time that we've birded here in May and the first time that we've sampled the riches of "Manchuria" in the n.e. provinces of Jilin and Inner Mongolia. I co-led one of those past tours here with Jesper, but this seemed, to me, to be a completely different tour given the season (those past tours were in September) and the coverage (the inclusion of Manchuria, and the addition of more sites on the Tibetan Plateau in Qinghai Province). There was some overlap this year in coverage with what Jesper and I did on my last tour here - some of the areas around Xining, Koko Nor (Qinghai Lake), Rubber Mountain, Chaka - but I was really impressed with the new bits that were added and I thought that it all made for a wonderful, bird-filled tour of this huge, diverse country. We started this year's trip off with a short visit to Wild Duck Lake near Beijing before flying north to Ulanhot in Jilin Province - what was once known widely as Manchuria. -

AERC TAC 2014 Recommendations Final Version July 2015 (Pdf)
AERC TAC’s TAXONOMIC RECOMMENDATIONS 2015 report Citation: Crochet P.-A., Barthel P.H., Bauer H.-G., van den Berg A.B., Bezzel E., Collinson J.M., Dubois P.J., Fromholtz J., Helbig A.J. †, Jiguet F., Jirle E., Knox A.G., Kirwan G., Lagerqvist M., Le Maréchal P., van Loon A.J., Päckert M., Parkin D.T., Pons J.-M., Raty L., Roselaar C.S., Sangster G., Steinheimer F.D., Svensson L., Tyrberg T., Votier S.C., Yésou P. (2015) AERC TAC's taxonomic recommendations: 2015 report. Available online at http://www.aerc.eu/tac.html. Published 15th July 2015. Introduction This document constitutes the official 2015 AERC TAC recommendations for species-level systematics and nomenclature of Western Palearctic birds. For full information on the TAC and its history, please refer to the documents on the AERC web page www.aerc.eu, including the minutes of the AERC meetings http://www.aerc.eu/minutes.html and the TAC pages www.aerc.eu/tac.html. The format of this document follows the previous TAC recommendations (Crochet et al. 2010), which see for details. The TAC has five members: Taxonomic Sub-committee of the British Ornithologists’ Union Records Committee (BOURC-TSC, UK), Commission de l’Avifaune Française (CAF, France), Swedish Taxonomic Committee (STC, Sweden), Commissie Systematiek Nederlandse Avifauna (CSNA, Netherlands) and Kommission Artenliste der Vögel Deutschlands der Deutschen Ornithologen-Gesellschaft (German TC, Germany). As decided previously, systematic changes are based on decisions published by, or directly passed to the TAC chairman, by these taxonomic committees (TCs). In this document, the support for each case is given as yes / no / not addressed. -

April 2019 (No.72) Reg.Charity No.508850
Teesmouth Bird Club Newsletter April 2019 (No.72) Reg.Charity No.508850 CONTENTS 1 Monthly Summaries MONTHLY SUMMARIES 3 Local Outings Martin Blick reviews the birding highlights of the November to February period. 4 Where are Guisboroughs Swifts NOVEMBER 2018 4 Titbits November was also rather exciting, rare and interesting birds turning up on almost every 6 A Book Worth Reading day of the month. 7 Hurworth Burn Reservoir Three Bearded Tits on Seaton Common on 1st were likely to have been the same three 8 Chopping and Changing noted by the Long Drag in Oct, a singing male Cetti’s Warbler was at Bowesfield from 10 Learning with Dinosaurs 2nd, a ringtail Hen Harrier came in off the sea at Hartlepool on 3rd, and a fourth Cetti’s 11 BTO News Warbler was heard at Haverton Hole from 5th. 13 Treasurers Report On that same day a White-tailed Eagle was found a mile or two South of the Cleveland 15 Report from the Chair boundary near Sleddale, but, despite numerous birders watching it for the next 17 Wetland Bird Survey three days, it did not venture into Cleveland. A Slavonian Grebe was on Bran Sands from 19 Club Membership 6th, a Pallid Swift was reported at North Gare on 7th and two at South Gare on the same 21 TBC Publications day, and three White-fronted Geese were at Scaling Dam on 9th. 22 TBC Clothing Sunday 11th November was Remembrance Day and the 100th anniversary of the signing of the treaty that ended the First World War. -

Sichuan, China Tour Report 2019
Pere David’s Owl (Craig Robson) SICHUAN 19 MAY – 7 JUNE 2019 LEADERS: CRAIG ROBSON The latest Birdquest tour to Sichuan Province in central China, once again proved to be a marvelous experience, with some really memorable birds and other wildlife. A total of 331 species of bird were recorded, including a large number of Sino-Himalayan specialties and Birdquest ‘diamond’ birds. Some of this year’s highlights were 15 species of Galliform, including Temminck’s Tragopan, Chinese Monal, White Eared, Blue Eared, Golden and Lady Amherst’s Pheasants, Black-necked Crane, Brown-cheeked Rail, Pere 1 BirdQuest Tour Report: Sichuan 2019 www.birdquest-tours.com David’s Owl, Saker Falcon, Tiger Shrike, Sichuan Jay, Collared Crow, Fire-capped, White-browed, Pere David’s, Sichuan, and Ground Tits, Tibetan Lark, Chinese Cupwing, Sooty Bushtit, Crested Tit-Warbler, Sichuan Bush Warbler, 19 species of Phylloscopus warbler, 28 species of ‘babbler’, including Golden- fronted, Chinese, Spectacled and Grey-hooded Fulvettas, Snowy-cheeked, Barred, Giant and Red-winged Laughingthrushes, Chinese Hwamei, Emei Shan Liocichla, and Rufous-tailed Babbler, 8 species of parrotbill, including Great, Spectacled, Ashy-throated, Grey-hooded, and Golden, Chinese and Przevalski’s Nuthatches, Wallcreeper, Kessler’s and Chinese Thrushes, Grandala, Fujian Niltava, Zappey’s Flycatcher, Chinese and Siberian Rubythroats, Firethroat, Blue-fronted Robin, Fork-tailed Sunbird, Black-winged, White- rumped and Red-necked Snowfinches, Robin and Brown Accentors, Przevalski’s Finch, Chinese Grosbeak, Pink-rumped, Sharpe’s, Dark-rumped, Three-banded, Chinese White-browed and Red-fronted Rosefinches, and Pine and Slaty Buntings. Our mammal tally was also high, with our total of 23 species including Grey Wolf, Tibetan Fox, two superb Red Pandas, Altai Weasel, Tufted Deer, Takin, Chinese Goral, Bharal and Tibetan Gazelle. -

Global Environment Facility (GEF) Operations
Conservation and sustainable use of biodiversity: Strengthening network of protected areas through advanced governance and management Part I: Project Information GEF ID 10113 Project Type FSP Type of Trust Fund GET CBIT/NGI CBIT NGI Project Title Conservation and sustainable use of biodiversity: Strengthening network of protected areas through advanced governance and management Countries Azerbaijan Agency(ies) FAO Other Executing Partner(s) Ministry of Environment and Natural Resources Executing Partner Type Government GEF Focal Area Biodiversity Taxonomy Focal Areas, Biodiversity, Tourism, Mainstreaming, Forestry - Including HCVF and REDD+, Agriculture and agrobiodiversity, Productive Landscapes, Protected Areas and Landscapes, Community Based Natural Resource Mngt, Terrestrial Protected Areas, Strengthen institutional capacity and decision-making, Influencing models, Demonstrate innovative approache, Individuals/Entrepreneurs, Private Sector, Stakeholders, Partnership, Type of Engagement, Strategic Communications, Communications, Local Communities, Beneficiaries, Community Based Organization, Civil Society, Academia, Gender-sensitive indicators, Gender Mainstreaming, Gender Equality, Women groups, Sex-disaggregated indicators, Access and control over natural resources, Gender results areas, Capacity Development, Participation and leadership, Capacity, Knowledge and Research, Field Visit, Knowledge Exchange, South-South, Peer-to-Peer Rio Markers Climate Change Mitigation Climate Change Mitigation 1 Climate Change Adaptation Climate Change Adaptation 1 Submission Date 10/5/2018 Expected Implementation Start 9/1/2021 Expected Completion Date 8/31/2026 Duration 60In Months Agency Fee($) 250,774.00 A. FOCAL/NON-FOCAL AREA ELEMENTS Objectives/Programs Focal Area Outcomes Trust GEF Co-Fin Fund Amount($) Amount($) BD-2-7 Address direct drivers to GET 1,000,000.00 3,500,000.00 protect habitats and species and improvie financial sustainability, effective management and ecosystem coverage of the global protected area estate.