Population Genetics and Phylogeography of the Pygmy Nuthatch in Southern California
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

FIELD GUIDES BIRDING TOURS: China: Manchuria & the Tibetan Plateau 2013
Field Guides Tour Report China: Manchuria & the Tibetan Plateau 2013 May 4, 2013 to May 25, 2013 Dave Stejskal & Jesper Hornskov For our tour description, itinerary, past triplists, dates, fees, and more, please VISIT OUR TOUR PAGE. Of the five crane species we saw on the tour, the Red-crowned Crane is probably the rarest in China, though they still are fairly common in Japan. (Photo by guide Dave Stejskal) Although this was Field Guides' third tour to China with co-leader Jesper Hornskov as our host, this was the first time that we've birded here in May and the first time that we've sampled the riches of "Manchuria" in the n.e. provinces of Jilin and Inner Mongolia. I co-led one of those past tours here with Jesper, but this seemed, to me, to be a completely different tour given the season (those past tours were in September) and the coverage (the inclusion of Manchuria, and the addition of more sites on the Tibetan Plateau in Qinghai Province). There was some overlap this year in coverage with what Jesper and I did on my last tour here - some of the areas around Xining, Koko Nor (Qinghai Lake), Rubber Mountain, Chaka - but I was really impressed with the new bits that were added and I thought that it all made for a wonderful, bird-filled tour of this huge, diverse country. We started this year's trip off with a short visit to Wild Duck Lake near Beijing before flying north to Ulanhot in Jilin Province - what was once known widely as Manchuria. -

Manchuria & the Tibetan Plateau 2014
Field Guides Tour Report China: Manchuria & the Tibetan Plateau 2014 May 5, 2014 to May 26, 2014 Dave Stejskal & Jesper Hornskov For our tour description, itinerary, past triplists, dates, fees, and more, please VISIT OUR TOUR PAGE. This was only the second time that Field Guides has offered this itinerary co-led by China expert Jesper Hornskov, and it was a roaring success again! The first half of our tour was spent in the rich wetlands and grassland habitats of northeastern China -- the region formerly known as Manchuria -- before we jetted our way to the west and the high elevations of Qinghai Province. We had a bit of a hiccup at the start of the tour, but we certainly made the best of it. In short order, after hearing the bad news about our flight, we had a bus and were out birding north of Beijing looking at some real gems, as well as getting a look at a section of the renowned Great Wall. Thanks to all for your flexibility and patience! Then we were off to Manchuria the next morning, making stops at three important reserves during our time there. Momoge Reserve, our first destination, had some water level issues that developed through the spring, but we were still able to locate our prize there, thanks to the scouting efforts of Mr. Li. Our first afternoon in the area found us staring through the scopes at nearly 300 Critically Endangered Siberian Cranes on their way north to the breeding grounds. Fantastic!! The next morning, we enjoyed leisurely looks at the Endangered Rufous-backed (Jankovski's) Bunting at Tumuji Reserve (hardly a reserve at all!). -

Improved Sampling at the Subspecies Level Solves a Taxonomic Dilemma Â
Molecular Phylogenetics and Evolution 107 (2017) 538–550 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev Improved sampling at the subspecies level solves a taxonomic dilemma – A case study of two enigmatic Chinese tit species (Aves, Passeriformes, Paridae, Poecile) Christian Tritsch a, Jochen Martens b, Yue-Hua Sun c, Wieland Heim d,e, Patrick Strutzenberger a, ⇑ Martin Päckert a, a Senckenberg Naturhistorische Sammlungen, Königsbrücker Landstrabe 159, D-01109 Dresden, Germany b Institut für Zoologie, Johannes Gutenberg-Universität, 55099 Mainz, Germany c Key Laboratory of Animal Ecology and Conservation, Institute of Zoology, Chinese Academy of Science, 100101 Beijing, China d Institut für Biochemie und Biologie, AG Tierökologie, Universität Potsdam, Maulbeerallee 1, 14469 Potsdam, Germany e Amur Bird Project, Roseggerstrabe 14, 14471 Potsdam, Germany article info abstract Article history: A recent full species-level phylogeny of tits, titmice and chickadees (Paridae) has placed the Chinese Received 1 July 2016 endemic black-bibbed tit (Poecile hypermelaenus) as the sister to the Palearctic willow tit (P. montanus). Revised 25 November 2016 Because this sister-group relationship is in striking disagreement with the traditional affiliation of P. Accepted 9 December 2016 hypermelaenus close to the marsh tit (P. palustris) we tested this phylogenetic hypothesis in a multi- Available online 11 December 2016 locus analysis with an extended taxon sampling including sixteen subspecies of willow tits and marsh tits. As a taxonomic reference we included type specimens in our analysis. The molecular genetic study Keywords: was complemented with an analysis of biometric data obtained from museum specimens. Our phyloge- Poecile hypermelaenus netic reconstructions, including a comparison of all GenBank data available for our target species, clearly Poecile weigoldicus Multi-locus phylogeny show that the genetic lineage previously identified as P. -

Far East Odyssey: Japan, Taiwan and SE China in Winter
Far East Odyssey: Japan, Taiwan and SE China in Winter Custom Tour: 9 February – 7 March 2015 Leader: Keith Barnes. All Photos taken on this tour Red-crowned Cranes doing their nuptial display in Hokkaido. There is little in the birding world to beat this amazing experience of a rare and stunning-looking bird in amazing wilderness, doing its thing! www.tropicalbirding.com Summary Perhaps one of the best parts of this job is designing super-cool custom itineraries that do extraordinary things. As far as I am aware no birding trip like this has been attempted before. It was essentially a mop-up trip of all the marquee bucket-list birds anyone could want to search for in the Far East during the winter. It ended up being a veritable smorgasbord of stonking great megas! The 270 odd species we saw may not sound like much, but considering that 60% of the time we were surrounded by snow and ice, it isn’t bad. Of course, if we had spent a lot of time list loading by looking for clients’ non-lifer alcids in Hokkaido, or non-target shorebirds elsewhere, we could have easily cracked 300. But this was not about list-loading, it was simply about getting clients exactly what they wanted out of their custom trip! I think this report is a great example of why Tropical Birding does this better than anyone else in the industry. We listen to our clients, and then execute what they want to get the perfect experience. Besides, this trip was about enjoying amazing experiences with quality birds rather than simply glimpsing rare species. -
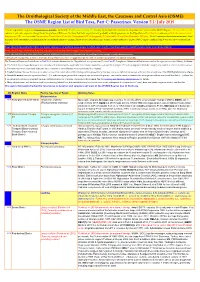
ORL 5.1 Passerines Final Draft01a.Xlsx
The Ornithological Society of the Middle East, the Caucasus and Central Asia (OSME) The OSME Region List of Bird Taxa, Part C: Passerines. Version 5.1: July 2019 A fuller explanation is given in Explanation of the ORL, but briefly, Bright green shading of a row (eg Syrian Ostrich) indicates former presence of a taxon in the OSME Region. Light gold shading in column A indicates sequence change from the previous ORL issue. For taxa that have unproven and probably unlikely presence, see the Hypothetical List. Red font indicates added information since the previous ORL version or the Conservation Threat Status (Critically Endangered = CE, Endangered = E, Vulnerable = V and Data Deficient = DD only). Not all synonyms have been examined. Serial numbers (SN) are merely an administrative convenience and may change. Please do not cite them in any formal correspondence or papers. NB: Compass cardinals (eg N = north, SE = southeast) are used. Rows shaded thus and with yellow text denote summaries of problem taxon groups in which some closely-related taxa may be of indeterminate status or are being studied. Rows shaded thus and with yellow text indicate recent or data-driven major conservation concerns. Rows shaded thus and with white text contain additional explanatory information on problem taxon groups as and when necessary. A broad dark orange line, as below, indicates the last taxon in a new or suggested species split, or where sspp are best considered separately. The Passerine Reference List follows as Part D, & includes References for Hypothetical non-passerines [List in Part E]. It explains Abbreviated References cited in the species accounts. -

The Tibetan Plateau & Migrants on China's East Coast
CHINA The Tibetan Plateau & Migrants on China’s East Coast 1st - 20 th October 2007 Henderson’s Ground-Jay and Tibetan Scenery All photos by David Shackelford Tour Leaders: Jesper Hornskov & David Shackelford Top 10 birds of the tour as voted by participants: 1. Black-necked Crane 2. Tibetan Snowcock 3. Red-crowned (Japanese) Crane 4. Wallcreeper 5. Mongolian (Henderson’s) Ground-Jay 6. Lammergeier 7. White-naped Crane 8. Pallas’s Sandgrouse 9. Crested Tit-Warbler 10. Przevalski’s Rosefinch RBT China & Tibet Trip Report October 2007 2 Group Photo on the Tibetan Plateau Tour Summary With the break of dawn in the misty mountains covered with broad spruce forest near Xining, provincial capital of northeast Tibet, we could see the silhouettes of literally hundreds of migrant Red-throated Thrushes spiraling through the air above us while the melodious songs of the endemic Pere-David’s and colorful Elliot’s Laughingthrushes serenaded us from the surrounding buckthorn scrub. The cold bite of winter above 2,000 meters kept us alert with a flurry of snowflakes at the higher altitudes as we sifted through noisy mixed flocks hosting both Chinese and spectacular Przevalski’s Nuthatches, Gray-headed Bullfinch, the little-known Gansu Leaf-Warbler, striking White-throated Redstart, and unbelievable sunlit views of the outrageous Crested Tit-Warbler. Already migration was in full swing as streams of raptors crested the mountain ridge and soared overhead including Greater Spotted Eagle, Oriental Honey-Buzzard, swift Eurasian Sparrowhawk, stately Northern Goshawk, and the recently recognized Himalayan Buzzard. Both Chestnut and the handsome Kessler’s Thrushes put on a show before we ventured to the nearby gravel slopes locating a flock of attractive Rosy Pipit, Godlewski’s Bunting, noisy but entertaining flocks of Red- billed Chough, and the local pale race of the Sinai Rosefinch.