Flow Cytometry – Data Compression
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Package 'Brotli'
Package ‘brotli’ May 13, 2018 Type Package Title A Compression Format Optimized for the Web Version 1.2 Description A lossless compressed data format that uses a combination of the LZ77 algorithm and Huffman coding. Brotli is similar in speed to deflate (gzip) but offers more dense compression. License MIT + file LICENSE URL https://tools.ietf.org/html/rfc7932 (spec) https://github.com/google/brotli#readme (upstream) http://github.com/jeroen/brotli#read (devel) BugReports http://github.com/jeroen/brotli/issues VignetteBuilder knitr, R.rsp Suggests spelling, knitr, R.rsp, microbenchmark, rmarkdown, ggplot2 RoxygenNote 6.0.1 Language en-US NeedsCompilation yes Author Jeroen Ooms [aut, cre] (<https://orcid.org/0000-0002-4035-0289>), Google, Inc [aut, cph] (Brotli C++ library) Maintainer Jeroen Ooms <[email protected]> Repository CRAN Date/Publication 2018-05-13 20:31:43 UTC R topics documented: brotli . .2 Index 4 1 2 brotli brotli Brotli Compression Description Brotli is a compression algorithm optimized for the web, in particular small text documents. Usage brotli_compress(buf, quality = 11, window = 22) brotli_decompress(buf) Arguments buf raw vector with data to compress/decompress quality value between 0 and 11 window log of window size Details Brotli decompression is at least as fast as for gzip while significantly improving the compression ratio. The price we pay is that compression is much slower than gzip. Brotli is therefore most effective for serving static content such as fonts and html pages. For binary (non-text) data, the compression ratio of Brotli usually does not beat bz2 or xz (lzma), however decompression for these algorithms is too slow for browsers in e.g. -
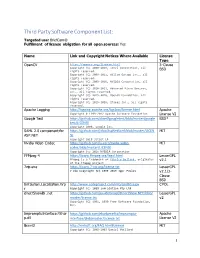
Third Party Software Component List: Targeted Use: Briefcam® Fulfillment of License Obligation for All Open Sources: Yes
Third Party Software Component List: Targeted use: BriefCam® Fulfillment of license obligation for all open sources: Yes Name Link and Copyright Notices Where Available License Type OpenCV https://opencv.org/license.html 3-Clause Copyright (C) 2000-2019, Intel Corporation, all BSD rights reserved. Copyright (C) 2009-2011, Willow Garage Inc., all rights reserved. Copyright (C) 2009-2016, NVIDIA Corporation, all rights reserved. Copyright (C) 2010-2013, Advanced Micro Devices, Inc., all rights reserved. Copyright (C) 2015-2016, OpenCV Foundation, all rights reserved. Copyright (C) 2015-2016, Itseez Inc., all rights reserved. Apache Logging http://logging.apache.org/log4cxx/license.html Apache Copyright © 1999-2012 Apache Software Foundation License V2 Google Test https://github.com/abseil/googletest/blob/master/google BSD* test/LICENSE Copyright 2008, Google Inc. SAML 2.0 component for https://github.com/jitbit/AspNetSaml/blob/master/LICEN MIT ASP.NET SE Copyright 2018 Jitbit LP Nvidia Video Codec https://github.com/lu-zero/nvidia-video- MIT codec/blob/master/LICENSE Copyright (c) 2016 NVIDIA Corporation FFMpeg 4 https://www.ffmpeg.org/legal.html LesserGPL FFmpeg is a trademark of Fabrice Bellard, originator v2.1 of the FFmpeg project 7zip.exe https://www.7-zip.org/license.txt LesserGPL 7-Zip Copyright (C) 1999-2019 Igor Pavlov v2.1/3- Clause BSD Infralution.Localization.Wp http://www.codeproject.com/info/cpol10.aspx CPOL f Copyright (C) 2018 Infralution Pty Ltd directShowlib .net https://github.com/pauldotknopf/DirectShow.NET/blob/ LesserGPL -

Metadefender Core V4.12.2
MetaDefender Core v4.12.2 © 2018 OPSWAT, Inc. All rights reserved. OPSWAT®, MetadefenderTM and the OPSWAT logo are trademarks of OPSWAT, Inc. All other trademarks, trade names, service marks, service names, and images mentioned and/or used herein belong to their respective owners. Table of Contents About This Guide 13 Key Features of Metadefender Core 14 1. Quick Start with Metadefender Core 15 1.1. Installation 15 Operating system invariant initial steps 15 Basic setup 16 1.1.1. Configuration wizard 16 1.2. License Activation 21 1.3. Scan Files with Metadefender Core 21 2. Installing or Upgrading Metadefender Core 22 2.1. Recommended System Requirements 22 System Requirements For Server 22 Browser Requirements for the Metadefender Core Management Console 24 2.2. Installing Metadefender 25 Installation 25 Installation notes 25 2.2.1. Installing Metadefender Core using command line 26 2.2.2. Installing Metadefender Core using the Install Wizard 27 2.3. Upgrading MetaDefender Core 27 Upgrading from MetaDefender Core 3.x 27 Upgrading from MetaDefender Core 4.x 28 2.4. Metadefender Core Licensing 28 2.4.1. Activating Metadefender Licenses 28 2.4.2. Checking Your Metadefender Core License 35 2.5. Performance and Load Estimation 36 What to know before reading the results: Some factors that affect performance 36 How test results are calculated 37 Test Reports 37 Performance Report - Multi-Scanning On Linux 37 Performance Report - Multi-Scanning On Windows 41 2.6. Special installation options 46 Use RAMDISK for the tempdirectory 46 3. Configuring Metadefender Core 50 3.1. Management Console 50 3.2. -

A Novel Coding Architecture for Multi-Line Lidar Point Clouds
This article has been accepted for inclusion in a future issue of this journal. Content is final as presented, with the exception of pagination. IEEE TRANSACTIONS ON INTELLIGENT TRANSPORTATION SYSTEMS 1 A Novel Coding Architecture for Multi-Line LiDAR Point Clouds Based on Clustering and Convolutional LSTM Network Xuebin Sun , Sukai Wang , Graduate Student Member, IEEE, and Ming Liu , Senior Member, IEEE Abstract— Light detection and ranging (LiDAR) plays an preservation of historical relics, 3D sensing for smart city, indispensable role in autonomous driving technologies, such as well as autonomous driving. Especially for autonomous as localization, map building, navigation and object avoidance. driving systems, LiDAR sensors play an indispensable role However, due to the vast amount of data, transmission and storage could become an important bottleneck. In this article, in a large number of key techniques, such as simultaneous we propose a novel compression architecture for multi-line localization and mapping (SLAM) [1], path planning [2], LiDAR point cloud sequences based on clustering and convolu- obstacle avoidance [3], and navigation. A point cloud consists tional long short-term memory (LSTM) networks. LiDAR point of a set of individual 3D points, in accordance with one or clouds are structured, which provides an opportunity to convert more attributes (color, reflectance, surface normal, etc). For the 3D data to 2D array, represented as range images. Thus, we cast the 3D point clouds compression as a range image instance, the Velodyne HDL-64E LiDAR sensor generates a sequence compression problem. Inspired by the high efficiency point cloud of up to 2.2 billion points per second, with a video coding (HEVC) algorithm, we design a novel compression range of up to 120 m. -

Compresso: Efficient Compression of Segmentation Data for Connectomics
Compresso: Efficient Compression of Segmentation Data For Connectomics Brian Matejek, Daniel Haehn, Fritz Lekschas, Michael Mitzenmacher, Hanspeter Pfister Harvard University, Cambridge, MA 02138, USA bmatejek,haehn,lekschas,michaelm,[email protected] Abstract. Recent advances in segmentation methods for connectomics and biomedical imaging produce very large datasets with labels that assign object classes to image pixels. The resulting label volumes are bigger than the raw image data and need compression for efficient stor- age and transfer. General-purpose compression methods are less effective because the label data consists of large low-frequency regions with struc- tured boundaries unlike natural image data. We present Compresso, a new compression scheme for label data that outperforms existing ap- proaches by using a sliding window to exploit redundancy across border regions in 2D and 3D. We compare our method to existing compression schemes and provide a detailed evaluation on eleven biomedical and im- age segmentation datasets. Our method provides a factor of 600-2200x compression for label volumes, with running times suitable for practice. Keywords: compression, encoding, segmentation, connectomics 1 Introduction Connectomics|reconstructing the wiring diagram of a mammalian brain at nanometer resolution|results in datasets at the scale of petabytes [21,8]. Ma- chine learning methods find cell membranes and create cell body labelings for every neuron [18,12,14] (Fig. 1). These segmentations are stored as label volumes that are typically encoded in 32 bits or 64 bits per voxel to support labeling of millions of different nerve cells (neurons). Storing such data is expensive and transferring the data is slow. To cut costs and delays, we need compression methods to reduce data sizes. -

Data Latency and Compression
Data Latency and Compression Joseph M. Steim1, Edelvays N. Spassov2 1Quanterra Inc., 2Kinemetrics, Inc. Abstract DIscussion More Compression Because of interest in the capability of digital seismic data systems to provide low-latency data for “Early Compression and Latency related to Origin Queuing[5] Lossless Lempel-Ziv (LZ) compression methods (e.g. gzip, 7zip, and more recently “brotli” introduced by Warning” applications, we have examined the effect of data compression on the ability of systems to Plausible data compression methods reduce the volume (number of bits) required to represent each data Google for web-page compression) are often used for lossless compression of binary and text files. How deliver data with low latency, and on the efficiency of data storage and telemetry systems. Quanterra Q330 sample. Compression of data therefore always reduces the minimum required bandwidth (bits/sec) for does the best example of modern LZ methods compare with FDSN Level 1 and 2 for waveform data? systems are widely used in telemetered networks, and are considered in particular. transmission compared with transmission of uncompressed data. The volume required to transmit Segments of data representing up to one month of various levels of compressibility were compressed using compressed data is variable, depending on characteristics of the data. Higher levels of compression require Levels 1,2 and 3, and brotli using “quality” 1 and 6. The compression ratio (relative to the original 32 bits) Q330 data acquisition systems transmit data compressed in Steim2-format packets. Some studies have less minimum bandwidth and volume, and maximize the available unused (“free”) bandwidth for shows that in all cases the compression using L1,L2, or L3 is significantly greater than brotli. -
![Archive and Compressed [Edit]](https://docslib.b-cdn.net/cover/8796/archive-and-compressed-edit-1288796.webp)
Archive and Compressed [Edit]
Archive and compressed [edit] Main article: List of archive formats • .?Q? – files compressed by the SQ program • 7z – 7-Zip compressed file • AAC – Advanced Audio Coding • ace – ACE compressed file • ALZ – ALZip compressed file • APK – Applications installable on Android • AT3 – Sony's UMD Data compression • .bke – BackupEarth.com Data compression • ARC • ARJ – ARJ compressed file • BA – Scifer Archive (.ba), Scifer External Archive Type • big – Special file compression format used by Electronic Arts for compressing the data for many of EA's games • BIK (.bik) – Bink Video file. A video compression system developed by RAD Game Tools • BKF (.bkf) – Microsoft backup created by NTBACKUP.EXE • bzip2 – (.bz2) • bld - Skyscraper Simulator Building • c4 – JEDMICS image files, a DOD system • cab – Microsoft Cabinet • cals – JEDMICS image files, a DOD system • cpt/sea – Compact Pro (Macintosh) • DAA – Closed-format, Windows-only compressed disk image • deb – Debian Linux install package • DMG – an Apple compressed/encrypted format • DDZ – a file which can only be used by the "daydreamer engine" created by "fever-dreamer", a program similar to RAGS, it's mainly used to make somewhat short games. • DPE – Package of AVE documents made with Aquafadas digital publishing tools. • EEA – An encrypted CAB, ostensibly for protecting email attachments • .egg – Alzip Egg Edition compressed file • EGT (.egt) – EGT Universal Document also used to create compressed cabinet files replaces .ecab • ECAB (.ECAB, .ezip) – EGT Compressed Folder used in advanced systems to compress entire system folders, replaced by EGT Universal Document • ESS (.ess) – EGT SmartSense File, detects files compressed using the EGT compression system. • GHO (.gho, .ghs) – Norton Ghost • gzip (.gz) – Compressed file • IPG (.ipg) – Format in which Apple Inc. -

Parquet Data Format Performance
Parquet data format performance Jim Pivarski Princeton University { DIANA-HEP February 21, 2018 1 / 22 What is Parquet? 1974 HBOOK tabular rowwise FORTRAN first ntuples in HEP 1983 ZEBRA hierarchical rowwise FORTRAN event records in HEP 1989 PAW CWN tabular columnar FORTRAN faster ntuples in HEP 1995 ROOT hierarchical columnar C++ object persistence in HEP 2001 ProtoBuf hierarchical rowwise many Google's RPC protocol 2002 MonetDB tabular columnar database “first” columnar database 2005 C-Store tabular columnar database also early, became HP's Vertica 2007 Thrift hierarchical rowwise many Facebook's RPC protocol 2009 Avro hierarchical rowwise many Hadoop's object permanance and interchange format 2010 Dremel hierarchical columnar C++, Java Google's nested-object database (closed source), became BigQuery 2013 Parquet hierarchical columnar many open source object persistence, based on Google's Dremel paper 2016 Arrow hierarchical columnar many shared-memory object exchange 2 / 22 What is Parquet? 1974 HBOOK tabular rowwise FORTRAN first ntuples in HEP 1983 ZEBRA hierarchical rowwise FORTRAN event records in HEP 1989 PAW CWN tabular columnar FORTRAN faster ntuples in HEP 1995 ROOT hierarchical columnar C++ object persistence in HEP 2001 ProtoBuf hierarchical rowwise many Google's RPC protocol 2002 MonetDB tabular columnar database “first” columnar database 2005 C-Store tabular columnar database also early, became HP's Vertica 2007 Thrift hierarchical rowwise many Facebook's RPC protocol 2009 Avro hierarchical rowwise many Hadoop's object permanance and interchange format 2010 Dremel hierarchical columnar C++, Java Google's nested-object database (closed source), became BigQuery 2013 Parquet hierarchical columnar many open source object persistence, based on Google's Dremel paper 2016 Arrow hierarchical columnar many shared-memory object exchange 2 / 22 Developed independently to do the same thing Google Dremel authors claimed to be unaware of any precedents, so this is an example of convergent evolution. -

The Deep Learning Solutions on Lossless Compression Methods for Alleviating Data Load on Iot Nodes in Smart Cities
sensors Article The Deep Learning Solutions on Lossless Compression Methods for Alleviating Data Load on IoT Nodes in Smart Cities Ammar Nasif *, Zulaiha Ali Othman and Nor Samsiah Sani Center for Artificial Intelligence Technology (CAIT), Faculty of Information Science & Technology, University Kebangsaan Malaysia, Bangi 43600, Malaysia; [email protected] (Z.A.O.); [email protected] (N.S.S.) * Correspondence: [email protected] Abstract: Networking is crucial for smart city projects nowadays, as it offers an environment where people and things are connected. This paper presents a chronology of factors on the development of smart cities, including IoT technologies as network infrastructure. Increasing IoT nodes leads to increasing data flow, which is a potential source of failure for IoT networks. The biggest challenge of IoT networks is that the IoT may have insufficient memory to handle all transaction data within the IoT network. We aim in this paper to propose a potential compression method for reducing IoT network data traffic. Therefore, we investigate various lossless compression algorithms, such as entropy or dictionary-based algorithms, and general compression methods to determine which algorithm or method adheres to the IoT specifications. Furthermore, this study conducts compression experiments using entropy (Huffman, Adaptive Huffman) and Dictionary (LZ77, LZ78) as well as five different types of datasets of the IoT data traffic. Though the above algorithms can alleviate the IoT data traffic, adaptive Huffman gave the best compression algorithm. Therefore, in this paper, Citation: Nasif, A.; Othman, Z.A.; we aim to propose a conceptual compression method for IoT data traffic by improving an adaptive Sani, N.S. -

Key-Based Self-Driven Compression in Columnar Binary JSON
Otto von Guericke University of Magdeburg Department of Computer Science Master's Thesis Key-Based Self-Driven Compression in Columnar Binary JSON Author: Oskar Kirmis November 4, 2019 Advisors: Prof. Dr. rer. nat. habil. Gunter Saake M. Sc. Marcus Pinnecke Institute for Technical and Business Information Systems / Database Research Group Kirmis, Oskar: Key-Based Self-Driven Compression in Columnar Binary JSON Master's Thesis, Otto von Guericke University of Magdeburg, 2019 Abstract A large part of the data that is available today in organizations or publicly is provided in semi-structured form. To perform analytical tasks on these { mostly read-only { semi-structured datasets, Carbon archives were developed as a column-oriented storage format. Its main focus is to allow cache-efficient access to fields across records. As many semi-structured datasets mainly consist of string data and the denormalization introduces redundancy, a lot of storage space is required. However, in Carbon archives { besides a deduplication of strings { there is currently no compression implemented. The goal of this thesis is to discuss, implement and evaluate suitable compression tech- niques to reduce the amount of storage required and to speed up analytical queries on Carbon archives. Therefore, a compressor is implemented that can be configured to apply a combination of up to three different compression algorithms to the string data of Carbon archives. This compressor can be applied with a different configuration per column (per JSON object key). To find suitable combinations of compression algo- rithms for each column, one manual and two self-driven approaches are implemented and evaluated. On a set of ten publicly available semi-structured datasets of different kinds and sizes, the string data can be compressed down to about 53% on average, reducing the whole datasets' size by 20%. -

Forcepoint DLP Supported File Formats and Size Limits
Forcepoint DLP Supported File Formats and Size Limits Supported File Formats and Size Limits | Forcepoint DLP | v8.8.1 This article provides a list of the file formats that can be analyzed by Forcepoint DLP, file formats from which content and meta data can be extracted, and the file size limits for network, endpoint, and discovery functions. See: ● Supported File Formats ● File Size Limits © 2021 Forcepoint LLC Supported File Formats Supported File Formats and Size Limits | Forcepoint DLP | v8.8.1 The following tables lists the file formats supported by Forcepoint DLP. File formats are in alphabetical order by format group. ● Archive For mats, page 3 ● Backup Formats, page 7 ● Business Intelligence (BI) and Analysis Formats, page 8 ● Computer-Aided Design Formats, page 9 ● Cryptography Formats, page 12 ● Database Formats, page 14 ● Desktop publishing formats, page 16 ● eBook/Audio book formats, page 17 ● Executable formats, page 18 ● Font formats, page 20 ● Graphics formats - general, page 21 ● Graphics formats - vector graphics, page 26 ● Library formats, page 29 ● Log formats, page 30 ● Mail formats, page 31 ● Multimedia formats, page 32 ● Object formats, page 37 ● Presentation formats, page 38 ● Project management formats, page 40 ● Spreadsheet formats, page 41 ● Text and markup formats, page 43 ● Word processing formats, page 45 ● Miscellaneous formats, page 53 Supported file formats are added and updated frequently. Key to support tables Symbol Description Y The format is supported N The format is not supported P Partial metadata -

Sequence Analysis Data-Dependent Bucketing Improves Reference-Free Compression of Sequencing Reads Rob Patro1 and Carl Kingsford2,*
Bioinformatics, 31(17), 2015, 2770–2777 doi: 10.1093/bioinformatics/btv248 Advance Access Publication Date: 24 April 2015 Original Paper Sequence analysis Data-dependent bucketing improves reference-free compression of sequencing reads Rob Patro1 and Carl Kingsford2,* 1Department of Computer Science, Stony Brook University, Stony Brook, NY 11794-4400, USA and 2Department Computational Biology, School of Computer Science, Carnegie Mellon University, 5000 Forbes Ave., Pittsburgh, PA 15213, USA *To whom correspondence should be addressed. Associate Editor: Inanc Birol Received on November 16, 2014; revised on April 11, 2015; accepted on April 20, 2015 Abstract Motivation: The storage and transmission of high-throughput sequencing data consumes signifi- cant resources. As our capacity to produce such data continues to increase, this burden will only grow. One approach to reduce storage and transmission requirements is to compress this sequencing data. Results: We present a novel technique to boost the compression of sequencing that is based on the concept of bucketing similar reads so that they appear nearby in the file. We demonstrate that, by adopting a data-dependent bucketing scheme and employing a number of encoding ideas, we can achieve substantially better compression ratios than existing de novo sequence compression tools, including other bucketing and reordering schemes. Our method, Mince, achieves up to a 45% reduction in file sizes (28% on average) compared with existing state-of-the-art de novo com- pression schemes. Availability and implementation: Mince is written in Cþþ11, is open source and has been made available under the GPLv3 license. It is available at http://www.cs.cmu.edu/ckingsf/software/mince.