Isolated Congenital Asplenia
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Different Genetic Mechanisms Mediate Spontaneous Versus UVR-Induced
RESEARCH ARTICLE Different genetic mechanisms mediate spontaneous versus UVR-induced malignant melanoma Blake Ferguson1, Herlina Y Handoko1, Pamela Mukhopadhyay1, Arash Chitsazan1, Lois Balmer2,3, Grant Morahan2, Graeme J Walker1†* 1Drug Discovery Group, QIMR Berghofer Medical Research Institute, Herston, Australia; 2Centre for Diabetes Research, Harry Perkins Institute of Medical Research, Perth, Australia; 3School of Medical and Health Sciences, Edith Cowan University, Joondalup, Australia Abstract Genetic variation conferring resistance and susceptibility to carcinogen-induced tumorigenesis is frequently studied in mice. We have now turned this idea to melanoma using the collaborative cross (CC), a resource of mouse strains designed to discover genes for complex diseases. We studied melanoma-prone transgenic progeny across seventy CC genetic backgrounds. We mapped a strong quantitative trait locus for rapid onset spontaneous melanoma onset to Prkdc, a gene involved in detection and repair of DNA damage. In contrast, rapid onset UVR-induced melanoma was linked to the ribosomal subunit gene Rrp15. Ribosome biogenesis was upregulated in skin shortly after UVR exposure. Mechanistically, variation in the ‘usual suspects’ by which UVR may exacerbate melanoma, defective DNA repair, melanocyte proliferation, or inflammatory cell infiltration, did not explain melanoma susceptibility or resistance across the CC. *For correspondence: [email protected] Instead, events occurring soon after exposure, such as dysregulation of ribosome function, which alters many aspects of cellular metabolism, may be important. † Present address: Experimental DOI: https://doi.org/10.7554/eLife.42424.001 Dermatology Group, The University of Queensland Diamantina Institute, Woolloongabba, Australia Introduction Competing interests: The Cutaneous malignant melanoma (MM) is well known to be associated with high levels of sun expo- authors declare that no sure. -

Allele-Specific Expression of Ribosomal Protein Genes in Interspecific Hybrid Catfish
Allele-specific Expression of Ribosomal Protein Genes in Interspecific Hybrid Catfish by Ailu Chen A dissertation submitted to the Graduate Faculty of Auburn University in partial fulfillment of the requirements for the Degree of Doctor of Philosophy Auburn, Alabama August 1, 2015 Keywords: catfish, interspecific hybrids, allele-specific expression, ribosomal protein Copyright 2015 by Ailu Chen Approved by Zhanjiang Liu, Chair, Professor, School of Fisheries, Aquaculture and Aquatic Sciences Nannan Liu, Professor, Entomology and Plant Pathology Eric Peatman, Associate Professor, School of Fisheries, Aquaculture and Aquatic Sciences Aaron M. Rashotte, Associate Professor, Biological Sciences Abstract Interspecific hybridization results in a vast reservoir of allelic variations, which may potentially contribute to phenotypical enhancement in the hybrids. Whether the allelic variations are related to the downstream phenotypic differences of interspecific hybrid is still an open question. The recently developed genome-wide allele-specific approaches that harness high- throughput sequencing technology allow direct quantification of allelic variations and gene expression patterns. In this work, I investigated allele-specific expression (ASE) pattern using RNA-Seq datasets generated from interspecific catfish hybrids. The objective of the study is to determine the ASE genes and pathways in which they are involved. Specifically, my study investigated ASE-SNPs, ASE-genes, parent-of-origins of ASE allele and how ASE would possibly contribute to heterosis. My data showed that ASE was operating in the interspecific catfish system. Of the 66,251 and 177,841 SNPs identified from the datasets of the liver and gill, 5,420 (8.2%) and 13,390 (7.5%) SNPs were identified as significant ASE-SNPs, respectively. -

(ACIP) General Best Guidance for Immunization
8. Altered Immunocompetence Updates This section incorporates general content from the Infectious Diseases Society of America policy statement, 2013 IDSA Clinical Practice Guideline for Vaccination of the Immunocompromised Host (1), to which CDC provided input in November 2011. The evidence supporting this guidance is based on expert opinion and arrived at by consensus. General Principles Altered immunocompetence, a term often used synonymously with immunosuppression, immunodeficiency, and immunocompromise, can be classified as primary or secondary. Primary immunodeficiencies generally are inherited and include conditions defined by an inherent absence or quantitative deficiency of cellular, humoral, or both components that provide immunity. Examples include congenital immunodeficiency diseases such as X- linked agammaglobulinemia, SCID, and chronic granulomatous disease. Secondary immunodeficiency is acquired and is defined by loss or qualitative deficiency in cellular or humoral immune components that occurs as a result of a disease process or its therapy. Examples of secondary immunodeficiency include HIV infection, hematopoietic malignancies, treatment with radiation, and treatment with immunosuppressive drugs. The degree to which immunosuppressive drugs cause clinically significant immunodeficiency generally is dose related and varies by drug. Primary and secondary immunodeficiencies might include a combination of deficits in both cellular and humoral immunity. Certain conditions like asplenia and chronic renal disease also can cause altered immunocompetence. Determination of altered immunocompetence is important to the vaccine provider because incidence or severity of some vaccine-preventable diseases is higher in persons with altered immunocompetence; therefore, certain vaccines (e.g., inactivated influenza vaccine, pneumococcal vaccines) are recommended specifically for persons with these diseases (2,3). Administration of live vaccines might need to be deferred until immune function has improved. -

Anti-MRPS17 Monoclonal Antibody, Clone FQS23694 (DCABH-6110) This Product Is for Research Use Only and Is Not Intended for Diagnostic Use
Anti-MRPS17 monoclonal antibody, clone FQS23694 (DCABH-6110) This product is for research use only and is not intended for diagnostic use. PRODUCT INFORMATION Product Overview Rabbit monoclonal to MRPS17 Antigen Description Mammalian mitochondrial ribosomal proteins are encoded by nuclear genes and help in protein synthesis within the mitochondrion. Mitochondrial ribosomes (mitoribosomes) consist of a small 28S subunit and a large 39S subunit. They have an estimated 75% protein to rRNA composition compared to prokaryotic ribosomes, where this ratio is reversed. Another difference between mammalian mitoribosomes and prokaryotic ribosomes is that the latter contain a 5S rRNA. Among different species, the proteins comprising the mitoribosome differ greatly in sequence, and sometimes in biochemical properties, which prevents easy recognition by sequence homology. This gene encodes a 28S subunit protein that belongs to the ribosomal protein S17P family. The encoded protein is moderately conserved between human mitochondrial and prokaryotic ribosomal proteins. Pseudogenes corresponding to this gene are found on chromosomes 1p, 3p, 6q, 14p, 18q, and Xq. Immunogen Recombinant fragment within Human MRPS17. The exact sequence is proprietary.Database link: Q9Y2R5 Isotype IgG Source/Host Rabbit Species Reactivity Human Clone FQS23694 Purity Tissue culture supernatant Conjugate Unconjugated Applications IHC-P, WB, ICC/IF, IP Positive Control HeLa, HepG2 and U937 cell lysate; Human kidney and liver tissue; HeLa cells Format Liquid Size 100 μl Buffer pH: 7.2; Preservative: 0.01% Sodium azide; Constituents: 49% PBS, 0.05% BSA, 50% Glycerol 45-1 Ramsey Road, Shirley, NY 11967, USA Email: [email protected] Tel: 1-631-624-4882 Fax: 1-631-938-8221 1 © Creative Diagnostics All Rights Reserved Preservative 0.01% Sodium Azide Storage Store at +4°C short term (1-2 weeks). -

Supplementary File 2A Revised
Supplementary file 2A. Differentially expressed genes in aldosteronomas compared to all other samples, ranked according to statistical significance. Missing values were not allowed in aldosteronomas, but to a maximum of five in the other samples. Acc UGCluster Name Symbol log Fold Change P - Value Adj. P-Value B R99527 Hs.8162 Hypothetical protein MGC39372 MGC39372 2,17 6,3E-09 5,1E-05 10,2 AA398335 Hs.10414 Kelch domain containing 8A KLHDC8A 2,26 1,2E-08 5,1E-05 9,56 AA441933 Hs.519075 Leiomodin 1 (smooth muscle) LMOD1 2,33 1,3E-08 5,1E-05 9,54 AA630120 Hs.78781 Vascular endothelial growth factor B VEGFB 1,24 1,1E-07 2,9E-04 7,59 R07846 Data not found 3,71 1,2E-07 2,9E-04 7,49 W92795 Hs.434386 Hypothetical protein LOC201229 LOC201229 1,55 2,0E-07 4,0E-04 7,03 AA454564 Hs.323396 Family with sequence similarity 54, member B FAM54B 1,25 3,0E-07 5,2E-04 6,65 AA775249 Hs.513633 G protein-coupled receptor 56 GPR56 -1,63 4,3E-07 6,4E-04 6,33 AA012822 Hs.713814 Oxysterol bining protein OSBP 1,35 5,3E-07 7,1E-04 6,14 R45592 Hs.655271 Regulating synaptic membrane exocytosis 2 RIMS2 2,51 5,9E-07 7,1E-04 6,04 AA282936 Hs.240 M-phase phosphoprotein 1 MPHOSPH -1,40 8,1E-07 8,9E-04 5,74 N34945 Hs.234898 Acetyl-Coenzyme A carboxylase beta ACACB 0,87 9,7E-07 9,8E-04 5,58 R07322 Hs.464137 Acyl-Coenzyme A oxidase 1, palmitoyl ACOX1 0,82 1,3E-06 1,2E-03 5,35 R77144 Hs.488835 Transmembrane protein 120A TMEM120A 1,55 1,7E-06 1,4E-03 5,07 H68542 Hs.420009 Transcribed locus 1,07 1,7E-06 1,4E-03 5,06 AA410184 Hs.696454 PBX/knotted 1 homeobox 2 PKNOX2 1,78 2,0E-06 -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Micrornas Mediated Regulation of the Ribosomal Proteins and Its Consequences on the Global Translation of Proteins
cells Review microRNAs Mediated Regulation of the Ribosomal Proteins and Its Consequences on the Global Translation of Proteins Abu Musa Md Talimur Reza 1,2 and Yu-Guo Yuan 1,3,* 1 Jiangsu Co-Innovation Center of Prevention and Control of Important Animal Infectious Diseases and Zoonoses, College of Veterinary Medicine, Yangzhou University, Yangzhou 225009, China; [email protected] 2 Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Pawi´nskiego5a, 02-106 Warsaw, Poland 3 Jiangsu Key Laboratory of Zoonosis/Joint International Research Laboratory of Agriculture and Agri-Product Safety, The Ministry of Education of China, Yangzhou University, Yangzhou 225009, China * Correspondence: [email protected]; Tel.: +86-514-8797-9228 Abstract: Ribosomal proteins (RPs) are mostly derived from the energy-consuming enzyme families such as ATP-dependent RNA helicases, AAA-ATPases, GTPases and kinases, and are important structural components of the ribosome, which is a supramolecular ribonucleoprotein complex, composed of Ribosomal RNA (rRNA) and RPs, coordinates the translation and synthesis of proteins with the help of transfer RNA (tRNA) and other factors. Not all RPs are indispensable; in other words, the ribosome could be functional and could continue the translation of proteins instead of lacking in some of the RPs. However, the lack of many RPs could result in severe defects in the biogenesis of ribosomes, which could directly influence the overall translation processes and global expression of the proteins leading to the emergence of different diseases including cancer. While microRNAs (miRNAs) are small non-coding RNAs and one of the potent regulators of the post-transcriptional 0 gene expression, miRNAs regulate gene expression by targeting the 3 untranslated region and/or coding region of the messenger RNAs (mRNAs), and by interacting with the 50 untranslated region, Citation: Reza, A.M.M.T.; Yuan, Y.-G. -

Practice Parameter for the Diagnosis and Management of Primary Immunodeficiency
Practice parameter Practice parameter for the diagnosis and management of primary immunodeficiency Francisco A. Bonilla, MD, PhD, David A. Khan, MD, Zuhair K. Ballas, MD, Javier Chinen, MD, PhD, Michael M. Frank, MD, Joyce T. Hsu, MD, Michael Keller, MD, Lisa J. Kobrynski, MD, Hirsh D. Komarow, MD, Bruce Mazer, MD, Robert P. Nelson, Jr, MD, Jordan S. Orange, MD, PhD, John M. Routes, MD, William T. Shearer, MD, PhD, Ricardo U. Sorensen, MD, James W. Verbsky, MD, PhD, David I. Bernstein, MD, Joann Blessing-Moore, MD, David Lang, MD, Richard A. Nicklas, MD, John Oppenheimer, MD, Jay M. Portnoy, MD, Christopher R. Randolph, MD, Diane Schuller, MD, Sheldon L. Spector, MD, Stephen Tilles, MD, Dana Wallace, MD Chief Editor: Francisco A. Bonilla, MD, PhD Co-Editor: David A. Khan, MD Members of the Joint Task Force on Practice Parameters: David I. Bernstein, MD, Joann Blessing-Moore, MD, David Khan, MD, David Lang, MD, Richard A. Nicklas, MD, John Oppenheimer, MD, Jay M. Portnoy, MD, Christopher R. Randolph, MD, Diane Schuller, MD, Sheldon L. Spector, MD, Stephen Tilles, MD, Dana Wallace, MD Primary Immunodeficiency Workgroup: Chairman: Francisco A. Bonilla, MD, PhD Members: Zuhair K. Ballas, MD, Javier Chinen, MD, PhD, Michael M. Frank, MD, Joyce T. Hsu, MD, Michael Keller, MD, Lisa J. Kobrynski, MD, Hirsh D. Komarow, MD, Bruce Mazer, MD, Robert P. Nelson, Jr, MD, Jordan S. Orange, MD, PhD, John M. Routes, MD, William T. Shearer, MD, PhD, Ricardo U. Sorensen, MD, James W. Verbsky, MD, PhD GlaxoSmithKline, Merck, and Aerocrine; has received payment for lectures from Genentech/ These parameters were developed by the Joint Task Force on Practice Parameters, representing Novartis, GlaxoSmithKline, and Merck; and has received research support from Genentech/ the American Academy of Allergy, Asthma & Immunology; the American College of Novartis and Merck. -

Mitochondrial Translation and Its Impact on Protein Homeostasis And
Mitochondrial translation and its impact on protein homeostasis and aging Tamara Suhm Academic dissertation for the Degree of Doctor of Philosophy in Biochemistry at Stockholm University to be publicly defended on Friday 15 February 2019 at 09.00 in Magnélisalen, Kemiska övningslaboratoriet, Svante Arrhenius väg 16 B. Abstract Besides their famous role as powerhouse of the cell, mitochondria are also involved in many signaling processes and metabolism. Therefore, it is unsurprising that mitochondria are no isolated organelles but are in constant crosstalk with other parts of the cell. Due to the endosymbiotic origin of mitochondria, they still contain their own genome and gene expression machinery. The mitochondrial genome of yeast encodes eight proteins whereof seven are core subunits of the respiratory chain and ATP synthase. These subunits need to be assembled with subunits imported from the cytosol to ensure energy supply of the cell. Hence, coordination, timing and accuracy of mitochondrial gene expression is crucial for cellular energy production and homeostasis. Despite the central role of mitochondrial translation surprisingly little is known about the molecular mechanisms. In this work, I used baker’s yeast Saccharomyces cerevisiae to study different aspects of mitochondrial translation. Exploiting the unique possibility to make directed modifications in the mitochondrial genome of yeast, I established a mitochondrial encoded GFP reporter. This reporter allows monitoring of mitochondrial translation with different detection methods and enables more detailed studies focusing on timing and regulation of mitochondrial translation. Furthermore, employing insights gained from bacterial translation, we showed that mitochondrial translation efficiency directly impacts on protein homeostasis of the cytoplasm and lifespan by affecting stress handling. -

The Role of Human Ribosomal Proteins in the Maturation of Rrna and Ribosome Production
JOBNAME: RNA 14#9 2008 PAGE: 1 OUTPUT: Friday August 8 17:34:50 2008 csh/RNA/164293/rna11320 Downloaded from rnajournal.cshlp.org on September 27, 2021 - Published by Cold Spring Harbor Laboratory Press The role of human ribosomal proteins in the maturation of rRNA and ribosome production SARA ROBLEDO,1,3 RACHEL A. IDOL,1,3 DAN L. CRIMMINS,2 JACK H. LADENSON,2 PHILIP J. MASON,1,4 and MONICA BESSLER1,4 1Department of Internal Medicine, Division of Hematology, Washington University School of Medicine, St. Louis, Missouri 63110, USA 2Department of Pathology and Immunology, Division of Laboratory and Genomic Medicine, Washington University School of Medicine, St. Louis, Missouri 63110, USA ABSTRACT Production of ribosomes is a fundamental process that occurs in all dividing cells. It is a complex process consisting of the coordinated synthesis and assembly of four ribosomal RNAs (rRNA) with about 80 ribosomal proteins (r-proteins) involving more than 150 nonribosomal proteins and other factors. Diamond Blackfan anemia (DBA) is an inherited red cell aplasia caused by mutations in one of several r-proteins. How defects in r-proteins, essential for proliferation in all cells, lead to a human disease with a specific defect in red cell development is unknown. Here, we investigated the role of r-proteins in ribosome biogenesis in order to find out whether those mutated in DBA have any similarities. We depleted HeLa cells using siRNA for several individual r-proteins of the small (RPS6, RPS7, RPS15, RPS16, RPS17, RPS19, RPS24, RPS25, RPS28) or large subunit (RPL5, RPL7, RPL11, RPL14, RPL26, RPL35a) and studied the effect on rRNA processing and ribosome production. -

Asplenia Vaccination Guide
Stanford Health Care Vaccination Subcommitee Revision date 11/308/2018 Functional or Anatomical Asplenia Vaccine Guide I. PURPOSE To outline appropriate vaccines targeting encapsulated bacteria for functionally or anatomically asplenic patients. Routine vaccines that may also be indicated but not addressed here include influenza, Tdap, herpes zoster, HPV, MMR, and varicella.1,2,3 II. Background Functionally or anatomically asplenic patients should be vaccinated to decrease the risk of sepsis due to organisms such as Streptococcus pneumoniae, Haemophilus influenzae type B, and Neisseria meningitidis. Guidelines are based on CDC recommendations. For additional information, see https://www.cdc.gov/vaccines/schedules/hcp/imz/adult-conditions.html. III. Procedures/Guidelines1,2,3,6,7,8 The regimen consists of 4 vaccines initially, followed by repeat doses as specified: 1. Haemophilus b conjugate (Hib) vaccine (ACTHIB®) IM once if they have not previously received Hib vaccine 2. Pneumococcal conjugate 13-valent (PCV13) vaccine (PREVNAR 13®) IM once • 2nd dose: Pneumococcal polysaccharide 23-valent (PPSV23) vaccine (PNEUMOVAX 23®) SQ/IM once given ≥ 8 weeks later, then 3rd dose as PPSV23 > 5 years later.4 Note: The above is valid for those who have not received any pneumococcal vaccines previously, or those with unknown vaccination history. If already received prior doses of PPSV23: give PCV13 at least 1 year after last PPSV23 dose. 3. Meningococcal conjugate vaccine (MenACWY-CRM, MENVEO®) IM (repeat in ≥ 8 weeks, then every 5 years thereafter) 4. Meningococcal serogroup B vaccine (MenB, BEXSERO®) IM (repeat in ≥ 4 weeks) Timing of vaccination relative to splenectomy: 1. Should be given at least 14 days before splenectomy, if possible. -
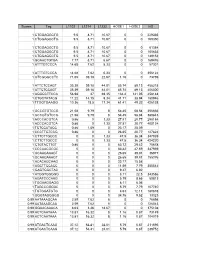
Scores Tag L1102 L1214 L1232 HOSE1 HOSE2 HS 1
Scores Tag L1102 L1214 L1232 HOSE1 HOSE2 HS 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 229335 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 169350 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 61384 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 105633 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 149152 1 GCAACTGTGA 7.77 8.71 6.67 0 0 169476 1 ATTTGTCCCA 14.68 7.62 5.33 0 0 57301 1 ATTTGTCCCA 14.68 7.62 5.33 0 0 356122 1 GTCGGGCCTC 71.65 39.18 22.67 1.16 0 73769 1 ATTCTCCAGT 35.39 39.18 44.01 85.74 89.13 458218 1 ATTCTCCAGT 35.39 39.18 44.01 85.74 89.13 406300 1 AGGGCTTCCA 56.98 37 69.35 134.4 141.05 458148 1 CTGCTATACG 11.22 14.15 9.34 41.71 38.94 180946 1 TTGGTGAAGG 10.36 18.5 17.34 61.41 49.32 426138 1 GCCGTGTCCG 21.58 9.79 8 54.45 58.84 356666 1 GCCGTGTCCG 21.58 9.79 8 54.45 58.84 380843 1 ACCCACGTCA 0.86 0 1.33 27.81 20.77 298184 1 ACCCACGTCA 0.86 0 1.33 27.81 20.77 400124 1 TCTCCATACC 0.86 1.09 0 23.17 25.09 1 CCCTTGTCCG 0.86 0 0 26.65 20.77 127824 1 CTTCTTGCCC 0 0 1.33 47.5 36.34 347939 1 CTTCTTGCCC 0 0 1.33 47.5 36.34 424220 1 CTGTACTTGT 0.86 0 0 63.72 29.42 75678 1 CCCAACGCGC 0 0 0 83.42 47.59 347939 1 GCAAGAAAGT 0 0 0 26.65 39.81 36977 1 GCAAGAAAGT 0 0 0 26.65 39.81 155376 1 ACACAGCAAG 0 0 0 23.17 15.58 1 AGCTTCCACC 0 0 0 11.59 7.79 355542 1 GAGTGGCTAC 0 0 0 9.27 6.92 1 ATGGTGGGGG 0 0 0 8.11 22.5 343586 1 AGATCCCAAG 0 0 0 5.79 8.65 50813 1 TGGAAGGAGG 0 0 0 8.11 6.06 1 TAGCCGGGAC 0 0 0 5.79 7.79 107740 1 TGTGGATGTG 0 0 0 4.63 12.11 180878 1 GGGTAGGGGG 0 0 0 34.76 9.52 13323 0.99 AATAAAGCAA 2.59 7.62 8 0 0 76698 0.99 AATAAAGCAA 2.59 7.62 8 0 0 126043 0.99 GGAACAAACA 8.63 3.26 18.67 0 0 375108