Transcriptional Decomposition Reveals Active Chromatin Architectures and Cell Specific Regulatory Interactions
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Análise Integrativa De Perfis Transcricionais De Pacientes Com
UNIVERSIDADE DE SÃO PAULO FACULDADE DE MEDICINA DE RIBEIRÃO PRETO PROGRAMA DE PÓS-GRADUAÇÃO EM GENÉTICA ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas Ribeirão Preto – 2012 ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas Tese apresentada à Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo para obtenção do título de Doutor em Ciências. Área de Concentração: Genética Orientador: Prof. Dr. Eduardo Antonio Donadi Co-orientador: Prof. Dr. Geraldo A. S. Passos Ribeirão Preto – 2012 AUTORIZO A REPRODUÇÃO E DIVULGAÇÃO TOTAL OU PARCIAL DESTE TRABALHO, POR QUALQUER MEIO CONVENCIONAL OU ELETRÔNICO, PARA FINS DE ESTUDO E PESQUISA, DESDE QUE CITADA A FONTE. FICHA CATALOGRÁFICA Evangelista, Adriane Feijó Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas. Ribeirão Preto, 2012 192p. Tese de Doutorado apresentada à Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo. Área de Concentração: Genética. Orientador: Donadi, Eduardo Antonio Co-orientador: Passos, Geraldo A. 1. Expressão gênica – microarrays 2. Análise bioinformática por module maps 3. Diabetes mellitus tipo 1 4. Diabetes mellitus tipo 2 5. Diabetes mellitus gestacional FOLHA DE APROVAÇÃO ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas. -
![Anti-CD150 / SLAM Antibody [SLAM.4] (FITC) (ARG42368)](https://docslib.b-cdn.net/cover/1644/anti-cd150-slam-antibody-slam-4-fitc-arg42368-1541644.webp)
Anti-CD150 / SLAM Antibody [SLAM.4] (FITC) (ARG42368)
Product datasheet [email protected] ARG42368 Package: 100 tests anti-CD150 / SLAM antibody [SLAM.4] (FITC) Store at: 4°C Summary Product Description FITC-conjugated Mouse Monoclonal antibody [SLAM.4] recognizes CD150 / SLAM Tested Reactivity Hu Tested Application FACS Specificity The mouse monoclonal antibody SLAM.4 recognizes an extracellular epitope of CD150, a cell surface molecule expressed on lymphocytes and involved in their activation. Host Mouse Clonality Monoclonal Clone SLAM.4 Isotype IgG1 Target Name CD150 / SLAM Antigen Species Human Immunogen Human CD150-transfected 300.19 cells. Conjugation FITC Alternate Names Signaling lymphocytic activation molecule; IPO-3; CD150; CDw150; CD antigen CD150; SLAM Application Instructions Application table Application Dilution FACS 4 µl / 100 µl of whole blood or 10^6 cells Application Note * The dilutions indicate recommended starting dilutions and the optimal dilutions or concentrations should be determined by the scientist. Calculated Mw 37 kDa Properties Form Liquid Purification Purified Buffer PBS and 15 mM Sodium azide. Preservative 15 mM Sodium azide Storage instruction Aliquot and store in the dark at 2-8°C. Keep protected from prolonged exposure to light. Avoid repeated freeze/thaw cycles. Suggest spin the vial prior to opening. The antibody solution should be gently mixed before use. Note For laboratory research only, not for drug, diagnostic or other use. www.arigobio.com 1/2 Bioinformation Gene Symbol SLAMF1 Gene Full Name signaling lymphocytic activation molecule family member 1 Function Self-ligand receptor of the signaling lymphocytic activation molecule (SLAM) family. SLAM receptors triggered by homo- or heterotypic cell-cell interactions are modulating the activation and differentiation of a wide variety of immune cells and thus are involved in the regulation and interconnection of both innate and adaptive immune response. -

Genetic and Functional Approaches to Understanding Autoimmune and Inflammatory Pathologies
University of Vermont ScholarWorks @ UVM Graduate College Dissertations and Theses Dissertations and Theses 2020 Genetic And Functional Approaches To Understanding Autoimmune And Inflammatory Pathologies Abbas Raza University of Vermont Follow this and additional works at: https://scholarworks.uvm.edu/graddis Part of the Genetics and Genomics Commons, Immunology and Infectious Disease Commons, and the Pathology Commons Recommended Citation Raza, Abbas, "Genetic And Functional Approaches To Understanding Autoimmune And Inflammatory Pathologies" (2020). Graduate College Dissertations and Theses. 1175. https://scholarworks.uvm.edu/graddis/1175 This Dissertation is brought to you for free and open access by the Dissertations and Theses at ScholarWorks @ UVM. It has been accepted for inclusion in Graduate College Dissertations and Theses by an authorized administrator of ScholarWorks @ UVM. For more information, please contact [email protected]. GENETIC AND FUNCTIONAL APPROACHES TO UNDERSTANDING AUTOIMMUNE AND INFLAMMATORY PATHOLOGIES A Dissertation Presented by Abbas Raza to The Faculty of the Graduate College of The University of Vermont In Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Specializing in Cellular, Molecular, and Biomedical Sciences January, 2020 Defense Date: August 30, 2019 Dissertation Examination Committee: Cory Teuscher, Ph.D., Advisor Jonathan Boyson, Ph.D., Chairperson Matthew Poynter, Ph.D. Ralph Budd, M.D. Dawei Li, Ph.D. Dimitry Krementsov, Ph.D. Cynthia J. Forehand, Ph.D., Dean of the Graduate College ABSTRACT Our understanding of genetic predisposition to inflammatory and autoimmune diseases has been enhanced by large scale quantitative trait loci (QTL) linkage mapping and genome-wide association studies (GWAS). However, the resolution and interpretation of QTL linkage mapping or GWAS findings are limited. -

Supplementary Table 1. List of Genes Up-Regulated in Abiraterone-Resistant Vcap Xenograft Samples PIK3IP1 Phosphoinositide-3-Kin
Supplementary Table 1. List of genes up-regulated in abiraterone-resistant VCaP xenograft samples PIK3IP1 phosphoinositide-3-kinase interacting protein 1 TMEM45A transmembrane protein 45A THBS1 thrombospondin 1 C7orf63 chromosome 7 open reading frame 63 OPTN optineurin FAM49A family with sequence similarity 49, member A APOL4 apolipoprotein L, 4 C17orf108|LOC201229 chromosome 17 open reading frame 108 | hypothetical protein LOC201229 SNORD94 small nucleolar RNA, C/D box 94 PCDHB11 protocadherin beta 11 RBM11 RNA binding motif protein 11 C6orf225 chromosome 6 open reading frame 225 KIAA1984|C9orf86|TMEM14 1 KIAA1984 | chromosome 9 open reading frame 86 | transmembrane protein 141 KIAA1107 TLR3 toll-like receptor 3 LPAR6 lysophosphatidic acid receptor 6 KIAA1683 GRB10 growth factor receptor-bound protein 10 TIMP2 TIMP metallopeptidase inhibitor 2 CCDC28A coiled-coil domain containing 28A FBXL2 F-box and leucine-rich repeat protein 2 NOV nephroblastoma overexpressed gene TSPAN31 tetraspanin 31 NR3C2 nuclear receptor subfamily 3, group C, member 2 DYNC2LI1 dynein, cytoplasmic 2, light intermediate chain 1 C15orf51 dynamin 1 pseudogene SAMD13 sterile alpha motif domain containing 13 RASSF6 Ras association (RalGDS/AF-6) domain family member 6 ZNF167 zinc finger protein 167 GATA2 GATA binding protein 2 NUDT7 nudix (nucleoside diphosphate linked moiety X)-type motif 7 DNAJC18 DnaJ (Hsp40) homolog, subfamily C, member 18 SNORA57 small nucleolar RNA, H/ACA box 57 CALCOCO1 calcium binding and coiled-coil domain 1 RLN2 relaxin 2 ING4 inhibitor of -

A Systems-Genetics Analyses of Complex Phenotypes
A systems-genetics analyses of complex phenotypes A thesis submitted to the University of Manchester for the degree of Doctor of Philosophy in the Faculty of Life Sciences 2015 David Ashbrook Table of contents Table of contents Table of contents ............................................................................................... 1 Tables and figures ........................................................................................... 10 General abstract ............................................................................................... 14 Declaration ....................................................................................................... 15 Copyright statement ........................................................................................ 15 Acknowledgements.......................................................................................... 16 Chapter 1: General introduction ...................................................................... 17 1.1 Overview................................................................................................... 18 1.2 Linkage, association and gene annotations .............................................. 20 1.3 ‘Big data’ and ‘omics’ ................................................................................ 22 1.4 Systems-genetics ..................................................................................... 24 1.5 Recombinant inbred (RI) lines and the BXD .............................................. 25 Figure 1.1: -

Rabbit Anti-SH2D1B/FITC Conjugated Antibody-SL19744R-FITC
SunLong Biotech Co.,LTD Tel: 0086-571- 56623320 Fax:0086-571- 56623318 E-mail:[email protected] www.sunlongbiotech.com Rabbit Anti-SH2D1B/FITC Conjugated antibody SL19744R-FITC Product Name: Anti-SH2D1B/FITC Chinese Name: FITC标记的EAT2蛋白抗体 EAT 2; EAT2; EWS/FLI1 activated transcript 2; SH2 domain containing 1B; SH2 Alias: domain containing molecule EAT2; SH2 domain containing protein 1B. Organism Species: Rabbit Clonality: Polyclonal React Species: Human,Mouse,Rat, ICC=1:50-200IF=1:50-200 Applications: not yet tested in other applications. optimal dilutions/concentrations should be determined by the end user. Molecular weight: 15kDa Form: Lyophilized or Liquid Concentration: 1mg/ml immunogen: KLH conjugated synthetic peptide derived from human SH2D1B Lsotype: IgG Purification: affinity purified by Protein A Storage Buffer: 0.01M TBS(pH7.4) with 1% BSA, 0.03% Proclin300 and 50% Glycerol. Storewww.sunlongbiotech.com at -20 °C for one year. Avoid repeated freeze/thaw cycles. The lyophilized antibody is stable at room temperature for at least one month and for greater than a year Storage: when kept at -20°C. When reconstituted in sterile pH 7.4 0.01M PBS or diluent of antibody the antibody is stable for at least two weeks at 2-4 °C. background: By binding phosphotyrosines through its free SRC (MIM 190090) homology-2 (SH2) domain, EAT2 regulates signal transduction through receptors expressed on the surface of antigen-presenting cells (Morra et al., 2001 [PubMed 11689425]).[supplied by Product Detail: OMIM, Mar 2008] Function: Plays a role in controlling signal transduction through at least four receptors, CD84, SLAMF1, LY9 and CD244, expressed on the surface of professional antigen-presenting cells. -

Meta-Analysis of Gene Expression in Individuals with Autism Spectrum Disorders
Meta-analysis of Gene Expression in Individuals with Autism Spectrum Disorders by Carolyn Lin Wei Ch’ng BSc., University of Michigan Ann Arbor, 2011 A THESIS SUBMITTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF Master of Science in THE FACULTY OF GRADUATE AND POSTDOCTORAL STUDIES (Bioinformatics) The University of British Columbia (Vancouver) August 2013 c Carolyn Lin Wei Ch’ng, 2013 Abstract Autism spectrum disorders (ASD) are clinically heterogeneous and biologically complex. State of the art genetics research has unveiled a large number of variants linked to ASD. But in general it remains unclear, what biological factors lead to changes in the brains of autistic individuals. We build on the premise that these heterogeneous genetic or genomic aberra- tions will converge towards a common impact downstream, which might be reflected in the transcriptomes of individuals with ASD. Similarly, a considerable number of transcriptome analyses have been performed in attempts to address this question, but their findings lack a clear consensus. As a result, each of these individual studies has not led to any significant advance in understanding the autistic phenotype as a whole. The goal of this research is to comprehensively re-evaluate these expression profiling studies by conducting a systematic meta-analysis. Here, we report a meta-analysis of over 1000 microarrays across twelve independent studies on expression changes in ASD compared to unaffected individuals, in blood and brain. We identified a number of genes that are consistently differentially expressed across studies of the brain, suggestive of effects on mitochondrial function. In blood, consistent changes were more difficult to identify, despite individual studies tending to exhibit larger effects than the brain studies. -
Download Validation Data
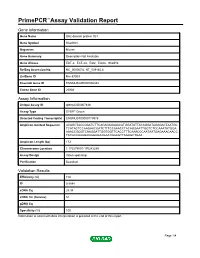
PrimePCR™Assay Validation Report Gene Information Gene Name SH2 domain protein 1B1 Gene Symbol Sh2d1b1 Organism Mouse Gene Summary Description Not Available Gene Aliases EAT-2, EAT-2A, Eat2, Eat2a, Sh2d1b RefSeq Accession No. NC_000067.6, NT_039185.8 UniGene ID Mm.57009 Ensembl Gene ID ENSMUSG00000096334 Entrez Gene ID 26904 Assay Information Unique Assay ID qMmuCID0007448 Assay Type SYBR® Green Detected Coding Transcript(s) ENSMUST00000179976 Amplicon Context Sequence ACAGCTACCGAATCTTCAGAGAGAAACATGGATATTACAGGATAGAGACTAATGC TCATACTCCAAGAACGATCTTTCCAAACCTACAGGAATTGGTCTCCAAATATGGA AAACCGGGTCAAGGATTGGTGGTTCACCTTTCAAACCCAATAATGAGAAACAACC TATGCCAAAGAGGGAGAAGAATGGAGTTAGAGCTGAA Amplicon Length (bp) 172 Chromosome Location 1:170279801-170283289 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 100 R2 0.9994 cDNA Cq 29.34 cDNA Tm (Celsius) 81 gDNA Cq Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 1/4 PrimePCR™Assay Validation Report Sh2d1b1, Mouse Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 2/4 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Mouse Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. Detected Coding Transcript(s) This is a list of the Ensembl transcript ID(s) that this assay will detect. Details for each transcript can be found on the Ensembl website at www.ensembl.org. -

SH2D1A (XLP 1D12) Rat Mab A
Revision 1 C 0 2 - t SH2D1A (XLP 1D12) Rat mAb a e r o t S Orders: 877-616-CELL (2355) [email protected] Support: 877-678-TECH (8324) 5 0 Web: [email protected] 8 www.cellsignal.com 2 # 3 Trask Lane Danvers Massachusetts 01923 USA For Research Use Only. Not For Use In Diagnostic Procedures. Applications: Reactivity: Sensitivity: MW (kDa): Source/Isotype: UniProt ID: Entrez-Gene Id: WB, F H Endogenous 14 Rat IgG2a O60880 4068 Product Usage Information Application Dilution Western Blotting 1:1000 Flow Cytometry 1:400 Storage Supplied in 10 mM sodium HEPES (pH 7.5), 150 mM NaCl, 100 µg/ml BSA, 50% glycerol and less than 0.02% sodium azide. Store at –20°C. Do not aliquot the antibody. Specificity / Sensitivity SH2D1A (XLP 1D12) Rat mAb detects endogenous levels of total SH2D1A protein. Species Reactivity: Human Source / Purification Monoclonal antibody is produced by immunizing animals with a recombinant full-length SH2D1A protein. Background SH2D1A and SH2D1B are small, adaptor proteins with a single SH2-domain that play important signal transduction roles mediated by the signaling lymphocytic activation molecule (SLAM) family receptors (1). SH2D1A (also called SAP or SLAM-associated protein) is frequently mutated in patients with X-linked lymphoproliferative disease (Duncan’s disease), which is characterized by extreme susceptibility to Epstein-Barr virus; approximately 50 different SH2D1A mutations have been reported to date (2-4). The single SH2D1B gene in humans (also called EAT-2 or Ewing's sarcoma's/FLI1-activated transcript 2) is present as a pair of duplicated EAT-2A and EAT-2B genes with identical genomic organization in mouse and rat (5,6). -

Content Based Search in Gene Expression Databases and a Meta-Analysis of Host Responses to Infection
Content Based Search in Gene Expression Databases and a Meta-analysis of Host Responses to Infection A Thesis Submitted to the Faculty of Drexel University by Francis X. Bell in partial fulfillment of the requirements for the degree of Doctor of Philosophy November 2015 c Copyright 2015 Francis X. Bell. All Rights Reserved. ii Acknowledgments I would like to acknowledge and thank my advisor, Dr. Ahmet Sacan. Without his advice, support, and patience I would not have been able to accomplish all that I have. I would also like to thank my committee members and the Biomed Faculty that have guided me. I would like to give a special thanks for the members of the bioinformatics lab, in particular the members of the Sacan lab: Rehman Qureshi, Daisy Heng Yang, April Chunyu Zhao, and Yiqian Zhou. Thank you for creating a pleasant and friendly environment in the lab. I give the members of my family my sincerest gratitude for all that they have done for me. I cannot begin to repay my parents for their sacrifices. I am eternally grateful for everything they have done. The support of my sisters and their encouragement gave me the strength to persevere to the end. iii Table of Contents LIST OF TABLES.......................................................................... vii LIST OF FIGURES ........................................................................ xiv ABSTRACT ................................................................................ xvii 1. A BRIEF INTRODUCTION TO GENE EXPRESSION............................. 1 1.1 Central Dogma of Molecular Biology........................................... 1 1.1.1 Basic Transfers .......................................................... 1 1.1.2 Uncommon Transfers ................................................... 3 1.2 Gene Expression ................................................................. 4 1.2.1 Estimating Gene Expression ............................................ 4 1.2.2 DNA Microarrays ...................................................... -

Differentially Expressed Genes in Aneurysm Tissue Compared With
On-line Table: Differentially expressed genes in aneurysm tissue compared with those in control tissue Fold False Discovery Direction of Gene Entrez Gene Name Function Change P Value Rate (q Value) Expression AADAC Arylacetamide deacetylase Positive regulation of triglyceride 4.46 1.33E-05 2.60E-04 Up-regulated catabolic process ABCA6 ATP-binding cassette, subfamily A (ABC1), Integral component of membrane 3.79 9.15E-14 8.88E-12 Up-regulated member 6 ABCC3 ATP-binding cassette, subfamily C (CFTR/MRP), ATPase activity, coupled to 6.63 1.21E-10 7.33E-09 Up-regulated member 3 transmembrane movement of substances ABI3 ABI family, member 3 Peptidyl-tyrosine phosphorylation 6.47 2.47E-05 4.56E-04 Up-regulated ACKR1 Atypical chemokine receptor 1 (Duffy blood G-protein–coupled receptor signaling 3.80 7.95E-10 4.18E-08 Up-regulated group) pathway ACKR2 Atypical chemokine receptor 2 G-protein–coupled receptor signaling 0.42 3.29E-04 4.41E-03 Down-regulated pathway ACSM1 Acyl-CoA synthetase medium-chain family Energy derivation by oxidation of 9.87 1.70E-08 6.52E-07 Up-regulated member 1 organic compounds ACTC1 Actin, ␣, cardiac muscle 1 Negative regulation of apoptotic 0.30 7.96E-06 1.65E-04 Down-regulated process ACTG2 Actin, ␥2, smooth muscle, enteric Blood microparticle 0.29 1.61E-16 2.36E-14 Down-regulated ADAM33 ADAM domain 33 Integral component of membrane 0.23 9.74E-09 3.95E-07 Down-regulated ADAM8 ADAM domain 8 Positive regulation of tumor necrosis 4.69 2.93E-04 4.01E-03 Up-regulated factor (ligand) superfamily member 11 production ADAMTS18 -
WO 2015/066640 Al 7 May 2015 (07.05.2015) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2015/066640 Al 7 May 2015 (07.05.2015) P O P C T (51) International Patent Classification: (81) Designated States (unless otherwise indicated, for every C12N 15/09 (2006.01) kind of national protection available): AE, AG, AL, AM, AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, (21) International Application Number: BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, PCT/US2014/063736 DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, (22) International Filing Date: HN, HR, HU, ID, IL, IN, IR, IS, JP, KE, KG, KN, KP, KR, 3 November 20 14 (03 .11.20 14) KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, (25) Filing Language: English PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, SC, (26) Publication Language: English SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (30) Priority Data: 61/898,928 1 November 2013 (01. 11.2013) US (84) Designated States (unless otherwise indicated, for every 61/898,932 1 November 2013 (01. 11.2013) US kind of regional protection available): ARIPO (BW, GH, GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, ST, SZ, (71) Applicant: THE RESEARCH INSTITUTE AT NA¬ TZ, UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TIONWIDE CHILDREN'S HOSPITAL [US/US]; 700 TJ, TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, Children's Drive, Columbus, Ohio 53305 (US).