Phylogeny and New Taxonomy of the Booted Eagles (Accipitriformes: Aquilinae)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Laws of Malaysia
LAWS OF MALAYSIA ONLINE VERSION OF UPDATED TEXT OF REPRINT Act 716 WILDLIFE CONSERVATION ACT 2010 As at 1 December 2014 2 WILDLIFE CONSERVATION ACT 2010 Date of Royal Assent … … 21 October 2010 Date of publication in the Gazette … … … 4 November 2010 Latest amendment made by P.U.(A)108/2014 which came into operation on ... ... ... ... … … … … 18 April 2014 3 LAWS OF MALAYSIA Act 716 WILDLIFE CONSERVATION ACT 2010 ARRANGEMENT OF SECTIONS PART I PRELIMINARY Section 1. Short title and commencement 2. Application 3. Interpretation PART II APPOINTMENT OF OFFICERS, ETC. 4. Appointment of officers, etc. 5. Delegation of powers 6. Power of Minister to give directions 7. Power of the Director General to issue orders 8. Carrying and use of arms PART III LICENSING PROVISIONS Chapter 1 Requirement for licence, etc. 9. Requirement for licence 4 Laws of Malaysia ACT 716 Section 10. Requirement for permit 11. Requirement for special permit Chapter 2 Application for licence, etc. 12. Application for licence, etc. 13. Additional information or document 14. Grant of licence, etc. 15. Power to impose additional conditions and to vary or revoke conditions 16. Validity of licence, etc. 17. Carrying or displaying licence, etc. 18. Change of particulars 19. Loss of licence, etc. 20. Replacement of licence, etc. 21. Assignment of licence, etc. 22. Return of licence, etc., upon expiry 23. Suspension or revocation of licence, etc. 24. Licence, etc., to be void 25. Appeals Chapter 3 Miscellaneous 26. Hunting by means of shooting 27. No licence during close season 28. Prerequisites to operate zoo, etc. 29. Prohibition of possessing, etc., snares 30. -

A Multi-Gene Phylogeny of Aquiline Eagles (Aves: Accipitriformes) Reveals Extensive Paraphyly at the Genus Level
Available online at www.sciencedirect.com MOLECULAR SCIENCE•NCE /W\/Q^DIRI DIRECT® PHYLOGENETICS AND EVOLUTION ELSEVIER Molecular Phylogenetics and Evolution 35 (2005) 147-164 www.elsevier.com/locate/ympev A multi-gene phylogeny of aquiline eagles (Aves: Accipitriformes) reveals extensive paraphyly at the genus level Andreas J. Helbig'^*, Annett Kocum'^, Ingrid Seibold^, Michael J. Braun^ '^ Institute of Zoology, University of Greifswald, Vogelwarte Hiddensee, D-18565 Kloster, Germany Department of Zoology, National Museum of Natural History, Smithsonian Institution, 4210 Silver Hill Rd., Suitland, MD 20746, USA Received 19 March 2004; revised 21 September 2004 Available online 24 December 2004 Abstract The phylogeny of the tribe Aquilini (eagles with fully feathered tarsi) was investigated using 4.2 kb of DNA sequence of one mito- chondrial (cyt b) and three nuclear loci (RAG-1 coding region, LDH intron 3, and adenylate-kinase intron 5). Phylogenetic signal was highly congruent and complementary between mtDNA and nuclear genes. In addition to single-nucleotide variation, shared deletions in nuclear introns supported one basal and two peripheral clades within the Aquilini. Monophyly of the Aquilini relative to other birds of prey was confirmed. However, all polytypic genera within the tribe, Spizaetus, Aquila, Hieraaetus, turned out to be non-monophyletic. Old World Spizaetus and Stephanoaetus together appear to be the sister group of the rest of the Aquilini. Spiza- stur melanoleucus and Oroaetus isidori axe nested among the New World Spizaetus species and should be merged with that genus. The Old World 'Spizaetus' species should be assigned to the genus Nisaetus (Hodgson, 1836). The sister species of the two spotted eagles (Aquila clanga and Aquila pomarina) is the African Long-crested Eagle (Lophaetus occipitalis). -

Chromosome Painting in Three Species of Buteoninae: a Cytogenetic Signature Reinforces the Monophyly of South American Species
Chromosome Painting in Three Species of Buteoninae: A Cytogenetic Signature Reinforces the Monophyly of South American Species Edivaldo Herculano C. de Oliveira1,2,3*, Marcella Mergulha˜o Tagliarini4, Michelly S. dos Santos5, Patricia C. M. O’Brien3, Malcolm A. Ferguson-Smith3 1 Laborato´rio de Cultura de Tecidos e Citogene´tica, SAMAM, Instituto Evandro Chagas, Ananindeua, PA, Brazil, 2 Faculdade de Cieˆncias Exatas e Naturais, ICEN, Universidade Federal do Para´, Bele´m, PA, Brazil, 3 Cambridge Resource Centre for Comparative Genomics, Cambridge, United Kingdom, 4 Programa de Po´s Graduac¸a˜oem Neurocieˆncias e Biologia Celular, ICB, Universidade Federal do Para´, Bele´m, PA, Brazil, 5 PIBIC – Universidade Federal do Para´, Bele´m, PA, Brazil Abstract Buteoninae (Falconiformes, Accipitridae) consist of the widely distributed genus Buteo, and several closely related species in a group called ‘‘sub-buteonine hawks’’, such as Buteogallus, Parabuteo, Asturina, Leucopternis and Busarellus, with unsolved phylogenetic relationships. Diploid number ranges between 2n = 66 and 2n = 68. Only one species, L. albicollis had its karyotype analyzed by molecular cytogenetics. The aim of this study was to present chromosomal analysis of three species of Buteoninae: Rupornis magnirostris, Asturina nitida and Buteogallus meridionallis using fluorescence in situ hybridization (FISH) experiments with telomeric and rDNA probes, as well as whole chromosome probes derived from Gallus gallus and Leucopternis albicollis. The three species analyzed herein showed similar karyotypes, with 2n = 68. Telomeric probes showed some interstitial telomeric sequences, which could be resulted by fusion processes occurred in the chromosomal evolution of the group, including the one found in the tassociation GGA1p/GGA6. -

Studies of Less Familiar Birds Ij2 Lesser Spotted Eagle B.-U
Studies of less familiar birds ij2 Lesser Spotted Eagle B.-U. Meyburg Plates 61-64. Of all European eagles, the Lesser Spotted Eagle Aquila pomarina has the smallest world breeding range, the nominate race being confined to eastern parts of Germany, Poland, eastern Czechoslovakia, Hun gary, Yugoslavia, Romania, Bulgaria, north-east Greece, western Turkey (Thrace), and the Soviet Union north to Leningrad and east to about 35°E. There are no published records of proved breeding in eastern Austria this century, though pairs from Hungary hunt over the land around Lake Neusiedl. There is no clear information on the current position in the Caucasus and the south Caspian lowlands. In Germany the breeding area stretches only to the north of Berlin, westwards beyond the rivers Oder and Neisse but stopping short of the Elbe. In 1969 there were 53 known broods in this area, and possibly a further nine (H. Weber in Glutz von Blotzheim et al. 1971). Even at the beginning of this century, the breeding range extended much further westwards, at least as far as the River Weser in Niedersachsen. A second subspecies, A. p. hastata, breeds in parts of India, in particular the Ganges Valley and West Bengal, and also in Bangla Desh. Hardly anything is known about this form, which appears to be rare. The first autumn plumage is said to be quite different from that of the nominate race, which has led some authors to treat it as a separate species. The Lesser Spotted Eagle presents a very difficult problem to field and museum ornithologists alike—the clear differentiation between it and the very closely related Spotted Eagle A. -

Effect of Environmental Elements on Migration Pattern of Eagles at Jorbeer Conservation Reserve, Bikaner, Rajasthan, India
Kataria et al RJLBPCS 2016 www.rjlbpcs.com Life Science Informatics Publications Original Research Article DOI - 10.26479/2016.0203.08 EFFECT OF ENVIRONMENTAL ELEMENTS ON MIGRATION PATTERN OF EAGLES AT JORBEER CONSERVATION RESERVE, BIKANER, RAJASTHAN, INDIA A.K. Kataria1*, N.Kataria2 and R.N.Kumawat3 1.Principal Investigator, Centre for excellence for use of space based technology in animal science, Rajasthan University of Veterinary and Animal Sciences, Bikaner-334001, Rajasthan, India. 2.Professor & Head, Department of Veterinary Physiology, College of Veterinary and Animal Science, Rajasthan University of Veterinary and Animal Sciences, Bikaner-334001, Rajasthan, India. 3. Deputy Forest Officer (Wildlife), Bikaner, Rajasthan ABSTRACT: The present endeavor was carried out to find out the effect of environmental elements on migration pattern of eagles at Jorbeer Conservation Reserve, Bikaner, Rajasthan, India (JCRBRI) during period from April 2015 to July 2016. The eagles studied were steppe eagle (Aquila nipalensis) and greater spotted eagle (Clanga clanga). The greater spotted eagle was seen only from November to February reiterating their winter migration to this reserve. Steppe eagle stayed at reserve during various months of the study year 2015 and 2016 when the THI values varied as 56.64-79.5. Recording of count of steppe eagle and greater spotted eagle besides real-time observation of environmental temperature and humidity signified in monitoring of eagle residence in the region. Result of the endeavor assisted in comprehending the absence of eagles in summer months during study periods. Residential period of steppe eagle was from November to April and of greater spotted eagle was from November to February only. -

Journal of Rese Arch in B Iolog Y
Journal of Research in Biology An International Scientific Research Journal Original Research Population density of Indian giant squirrel Ratufa indica centralis (Ryley, 1913) in Satpura National Park, Madhya Pradesh, India Authors: ABSTRACT: 1 Raju Lal Gurjar , 1* Amol .S. Kumbhar , Jyotirmay Jena1, 1 Information on population and distributional status of Indian giant squirrel Jaya Kumar Yogesh , Ratufa indica centralis is poorly known from central Indian hills. The species is Chittaranjan Dave1, endemic to India and widely distributed in Western Ghats, Eastern Ghats and Central Ramesh Pratap Singh2, India. In this study using line transect distance sampling we estimated population Ashok Mishra2. density of giant squirrel in Satpura Tiger Reserve (STR), which is a major biosphere Institution: reserve in central India that harbors wide variety of rare endemic and endangered 1. WWF - India, Nisha species. Density estimate with total effort of 276km line transect shows 5.5 (± 0.82) 2 Building, Near Forest squirrels/Km . This study provides first baseline information on ecological density Barrier, Katra, Mandla, estimate of Ratufa indica centralis in central Indian landscape. Reduction of Madhya Pradesh, India. anthropogenic pressure should be the first priority for park managers in Satpura Tiger reserve. 2. Field Director Office, Satpura Tiger Reserve, Hoshangabad, Madhya Pradesh, India. Corresponding author: Keywords: Amol S. Kumbhar Central Indian landscape, Distance sampling, density estimation, Ratufa indica centralis. Email Id: Article Citation: Raju Lal Gurjar, Amol S. Kumbhar, Jyotirmay Jena, Jaya Kumar Yogesh, Chittaranjan Dave, Ramesh Pratap Singh and Ashok Mishra. Population density of Indian giant squirrel Ratufa indica centralis (Ryley, 1913) in Journal of Research in Biology Research Journal of Satpura National Park, Madhya Pradesh, India. -

Bald Eagle Haliaeetus Leucocephalus
Wyoming Species Account Bald Eagle Haliaeetus leucocephalus REGULATORY STATUS USFWS: Delisted; Migratory Bird USFS R2: Sensitive USFS R4: Sensitive Wyoming BLM: Sensitive State of Wyoming: Protected Bird CONSERVATION RANKS USFWS: Bird of Conservation Concern WGFD: NSS3 (Bb), Tier II WYNDD: G5, S4B/S5N Wyoming Contribution: LOW IUCN: Least Concern PIF Continental Concern Score: 9 STATUS AND RANK COMMENTS Bald Eagle (Haliaeetus leucocephalus) is provided international protection under the Federal Migratory Bird Treaty Act of 1918, as amended 1. In 1940, Bald Eagle was provided protection under the Bald and Golden Eagle Protection Act 2. In 1966, the southern subspecies was listed as federally endangered under the Endangered Species Preservation Act; the entire population in the contiguous United States was listed as endangered in 1978 under the 1973 Endangered Species Act (ESA). A significant increase in numbers of nesting pairs, productivity, and distribution allowed Bald Eagle to be reclassified from Endangered to Threatened in 1995 under the ESA 3. Bald Eagle was delisted in 2007, and numbers are considered to be stable to increasing across its range 4. The species has been assigned different state conservation ranks by the Wyoming Natural Diversity Database for the breeding season and nonbreeding season because the abundance of the species is different between seasons. NATURAL HISTORY Taxonomy: Bald Eagle is a member of the family Accipitridae, which includes kites, eagles, harriers, and hawks 5. There are two subspecies of Bald Eagle; H. l. alascanus is found north of 40 degrees latitude across North America, including Wyoming, while H. l. leucocephalus is found south of 40 degrees latitude in the Gulf coast states 6. -

Birdingasia 23Cover
100 BirdingASIA 23 (2015): 100–101 NOTEBOOK Notes on the diet of the Black Eagle Ictinaetus malaiensis ZHU LEI, YANG XIAO-NONG, HAO GUANG, LIU TIAN-TIAN, DAI ZI-YUE & SUN YUE-HUA Introduction reappeared, one of them with what was clearly a The Black Eagle Ictinaetus malaiensis is a large large mammal dangling from its claws; it then flew diurnal raptor which inhabits mountain forests of due north before disappearing from view. Although tropical and subtropical Asia (Clark 1994). the observation only lasted about a minute, DZ-Y Although widespread in Asia, it is not well known obtained images (Plates 1 & 2) which enabled the and most of the relatively few studies have focused animal to be identified as a Red and White Giant on breeding biology (Zhu et al. 2014). Here we Flying Squirrel. The observation also suggests that present a note of our observations of the species at the pair might have engaged in cooperative hunting. Wawu Shan National Forestry Park (NFP), central Sichuan, China (29.650°N 102.933°E), including Notes on diet predation of a nocturnal Red and White Giant Flying Although the Black Eagle is widely described as Squirrel Petaurista alborufus, and review available feeding on rodents, snakes, lizards, pheasants and, information on the diet of the Black Eagle. particularly, eggs and nestlings of other birds (Ferguson-Lees & Christie 2001, Ali 2005, Robson Observations 2008) or regarded as a small mammal specialist On 29 April 2011 at 12h10 the authors in the course (Rasmussen & Anderton 2012), few studies have of their fieldwork were watching a pair of Black focused on its diet and feeding habits. -

Status of the Eastern Imperial Eagle (Aquila Heliaca) in the European Part of Turkey
ACTA ZOOLOGICA BULGARICA Acta zool. bulg., Suppl. 3, 2011: 87-93 Status of the Eastern Imperial Eagle (Aquila heliaca) in the European part of Turkey Dimitar A. Demerdzhiev1, Stoycho A. Stoychev2, Nikolay G. Terziev2, and Ivaylo D. Angelov2 1 31 Bulgaria Blv�., 4230 Asenovgra�, Bulgaria; E�mails: �emer�jiev@yahoo.�om; �_�emer�[email protected]; w��.bspb.org 2 Haskovo 6300, P.O.Box 130, Bulgaria; E�mails: stoy�hev.s@gmail.�om; w��.bspb.org; [email protected]; ivailoange� [email protected]; w��.bspb.org Abstract: This arti�le presents the results of the �rst more �etaile� stu�ying on the �istribution an� numbers of the Eastern Imperial Eagle (Аquila heliaca SA V I G NY 1809) population in the European part of Turkey. T�enty territories o��upie� by Imperial Eagle pairs, �istribute� in three �ifferent regions �ere �is�overe� �uring the perio� 2008�2009. The bree�ing population was estimate� at 30�50 pairs. The stu�y i�enti�e� t�o main habitat types typi�al of the Imperial Eagles in European Turkey – open hilly areas an� lo� mountain areas (up to 450 m a.s.l.) an� lo� relief plain areas (50�150 m a.s.l.). Poplar trees (Populus sp. L) were i�enti�e� as the most preferre� nesting substratum (44%), follo�e� by Oaks (Quercus sp. L) (40%). Bree�ing �ensity �as 1 pair/100 km2 in both habitat types. The shortest �istan�e bet�een t�o bree�ing pairs �as 5.8 km re�or�e� in plain areas in the Thra�e region. -

Iucn Red Data List Information on Species Listed On, and Covered by Cms Appendices
UNEP/CMS/ScC-SC4/Doc.8/Rev.1/Annex 1 ANNEX 1 IUCN RED DATA LIST INFORMATION ON SPECIES LISTED ON, AND COVERED BY CMS APPENDICES Content General Information ................................................................................................................................................................................................................................ 2 Species in Appendix I ............................................................................................................................................................................................................................... 3 Mammalia ............................................................................................................................................................................................................................................ 4 Aves ...................................................................................................................................................................................................................................................... 7 Reptilia ............................................................................................................................................................................................................................................... 12 Pisces ................................................................................................................................................................................................................................................. -

An Early Pleistocene Eagle from Nebraska
248 SHORT COMMUNICATIONS kunthii blossoms by Bombus queens occurred and depend on many factors. That hummingbirds and the workers were unable to secure nectar while positioned ancestor of P. kunthii co-existed may be assumed; within the floral tube, probably as much as lo-20 otherwise its adaptation to hummingbird pollination per cent more nectar was available to Bombus p&her would make little sense. Thus it is possible that P. and Bombus trinominatus populations during this kunthii could have undergone much of its development period due to the feeding activity of Diglossa. under selective pressure from hummingbirds; still, it is clear that Diglossa baritula has co-existed with DISCUSSION AND CONCLUSIONS hummingbirds throughout New World montane hab- Grant and Grant (1968) have proposed an explanation itats for some time and therefore an earlier and more for the reciprocal evolution of hummingbirds and the important role in the evolution of P. kunthii would plants upon which they feed. According to this inter- not be unexpected. This is not to suggest that exploita- pretation most hummingbird-pollinated flowers, espe- tion late in the evolutionary development of P. kunthii cially temperate species, have evolved from bee flowers would be insignificant. Even at present, given the (Grant 1961; Grant and Grant 1965). The process potential counter-selection pressures on P. kunthii involves an incipient stage during which a primitive from bees, the presence of Dglossa perforations un- hummingbird or progenitor already “preadapted” to doubtedly precludes a certain amount of bee pollina- feed on a particular bee flower (in the sense of tion which would probably otherwise occur, helping securing insects within the corolla, or nectar, or both), to maintain the selection pressures on P. -
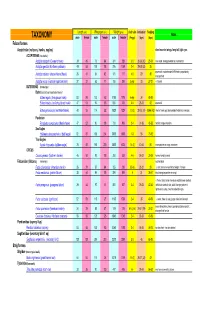
Avian Taxonomy
Length (cm) Wing span (cm) Weight (gms) cluch size incubation fledging Notes TAXONOMY male female male female male female (# eggs) (days) (days) Falconiformes Accipitridae (vultures, hawks, eagles) short rounded wings; long tail; light eyes ACCIPITRINAE (true hawks) Accipiter cooperii (Cooper's hawk) 39 45 73 84 341 528 3-5 30-36 (30) 25-34 crow sized; strongly banded tail; rounded tail Accipiter gentilis (Northern goshawk) 49 58 101 108 816 1059 2-4 28-38 (33) 35 square tail; most abundant NAM hawk; proportionaly 26 31 54 62 101 177 4-5 29 30 Accipiter striatus (sharp-shinned hawk) strongest foot Accipiter nisus (Eurasian sparrowhawk ) 37 37 62 77 150 290 5-Apr 33 27-31 Musket BUTEONINAE (broadwings) Buteo (buzzards or broad tailed hawks) Buteo regalis (ferruginous hawk) 53 59 132 143 1180 1578 6-Apr 34 45-50 Buteo lineatus (red-shouldered hawk) 47 53 96 105 550 700 3-4 28-33 42 square tail Buteo jamaicensis (red-tailed hawk) 48 55 114 122 1028 1224 1-3 (3) 28-35 (34) 42-46 (42) (Harlan' hawk spp); dark patagial featehres: immature; Parabuteo Parabuteo cuncincutus (Harris hawk) 47 52 90 108 710 890 2-4 31-36 45-50 reddish orange shoulders Sea Eagles Haliaeetus leucocephalus (bald eagle) 82 87 185 244 3000 6300 1-3 35 70-92 True Eagles Aquila chrysaetos (golden eagle) 78 82 185 220 3000 6125 1-4 (2) 40-45 50 white patches on wings: immature; CIRCUS Circus cyaneus (Northern harrier) 46 50 93 108 350 530 4-6 26-32 30-35 hovers; hunts by sound Falconidae (falcons) (longwings) notched beak Falco columbarius (American merlin) 26 29 57 64