Visualization and Biology: Fertile Ground for Collaboration
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
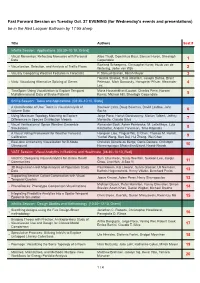
Tuesday's Evening Fast Forward Session
Fast Forward Session on Tuesday Oct. 27 EVENING (for Wednesday's events and presentations) be in the Red Lacquer Ballroom by 17:55 sharp Title Authors Seat # InfoVis Session: Applications [08:30–10:10, Grand] Visual Mementos: Reflecting Memories with Personal Alice Thudt, Dominikus Baur, Samuel Huron, Sheelagh - Data Carpendale 1 Roeland Scheepens, Christophe Hurter, Huub van de - Visualization, Selection, and Analysis of Traffic Flows Wetering, Jarke van Wijk 2 - Visually Comparing Weather Features in Forecasts P. Samuel Quinan, Miriah Meyer 3 Hendrik Strobelt, Bilal Alsallakh, Joseph Botros, Brant - Vials: Visualizing Alternative Splicing of Genes Peterson, Mark Borowsky, Hanspeter Pfister, Alexander 4 Lex TimeSpan: Using Visualization to Explore Temporal Mona Hosseinkhani Loorak, Charles Perin, Noreen - Multidimensional Data of Stroke Patients Kamal, Michael Hill, Sheelagh Carpendale 5 SciVis Session: Tasks and Applications [08:30–10:10, State] A Classification of User Tasks in Visual Analysis of Bireswar Laha, Doug Bowman, David Laidlaw, John - Volume Data Socha 6 Using Maximum Topology Matching to Explore Jorge Poco, Harish Doraiswamy, Marian Talbert, Jeffrey - Differences in Species Distribution Models Morisette, Claudio Silva 7 Visual Verification of Space Weather Ensemble Alexander Bock, Asher Pembroke, M. Leila Mays, Lutz - Simulations Rastaetter, Anders Ynnerman, Timo Ropinski 8 A Visual Voting Framework for Weather Forecast Hongsen Liao, Yingcai Wu, Li Chen, Thomas M. Hamill, - Calibration Yunhai Wang, Kan Dai, Hui Zhang, Wei -

Ways of Seeing Data: a Survey of Fields of Visualization
• Ways of seeing data: • A survey of fields of visualization • Gordon Kindlmann • [email protected] Part of the talk series “Show and Tell: Visualizing the Life of the Mind” • Nov 19, 2012 http://rcc.uchicago.edu/news/show_and_tell_abstracts.html 2012 Presidential Election REPUBLICAN DEMOCRAT http://www.npr.org/blogs/itsallpolitics/2012/11/01/163632378/a-campaign-map-morphed-by-money 2012 Presidential Election http://gizmodo.com/5960290/this-is-the-real-political-map-of-america-hint-we-are-not-that-divided 2012 Presidential Election http://www.npr.org/blogs/itsallpolitics/2012/11/01/163632378/a-campaign-map-morphed-by-money 2012 Presidential Election http://www.npr.org/blogs/itsallpolitics/2012/11/01/163632378/a-campaign-map-morphed-by-money 2012 Presidential Election http://www.npr.org/blogs/itsallpolitics/2012/11/01/163632378/a-campaign-map-morphed-by-money Clarifying distortions Tube map from 1908 http://www.20thcenturylondon.org.uk/beck-henry-harry http://briankerr.wordpress.com/2009/06/08/connections/ http://en.wikipedia.org/wiki/Harry_Beck Clarifying distortions http://www.20thcenturylondon.org.uk/beck-henry-harry Harry Beck 1933 http://briankerr.wordpress.com/2009/06/08/connections/ http://en.wikipedia.org/wiki/Harry_Beck Clarifying distortions Clarifying distortions Joachim Böttger, Ulrik Brandes, Oliver Deussen, Hendrik Ziezold, “Map Warping for the Annotation of Metro Maps” IEEE Computer Graphics and Applications, 28(5):56-65, 2008 Maps reflect conventions, choices, and priorities “A single map is but one of an indefinitely large -

Scientific Visualization
Report from Dagstuhl Seminar 11231 Scientific Visualization Edited by Min Chen1, Hans Hagen2, Charles D. Hansen3, and Arie Kaufman4 1 University of Oxford, GB, [email protected] 2 TU Kaiserslautern, DE, [email protected] 3 University of Utah, US, [email protected] 4 SUNY – Stony Brook, US, [email protected] Abstract This report documents the program and the outcomes of Dagstuhl Seminar 11231 “Scientific Visualization”. Seminar 05.–10. June, 2011 – www.dagstuhl.de/11231 1998 ACM Subject Classification I.3 Computer Graphics, I.4 Image Processing and Computer Vision, J.2 Physical Sciences and Engineering, J.3 Life and Medical Sciences Keywords and phrases Scientific Visualization, Biomedical Visualization, Integrated Multifield Visualization, Uncertainty Visualization, Scalable Visualization Digital Object Identifier 10.4230/DagRep.1.6.1 1 Executive Summary Min Chen Hans Hagen Charles D. Hansen Arie Kaufman License Creative Commons BY-NC-ND 3.0 Unported license © Min Chen, Hans Hagen, Charles D. Hansen, and Arie Kaufman Scientific Visualization (SV) is the transformation of abstract data, derived from observation or simulation, into readily comprehensible images, and has proven to play an indispensable part of the scientific discovery process in many fields of contemporary science. This seminar focused on the general field where applications influence basic research questions on one hand while basic research drives applications on the other. Reflecting the heterogeneous structure of Scientific Visualization and the currently unsolved problems in the field, this seminar dealt with key research problems and their solutions in the following subfields of scientific visualization: Biomedical Visualization: Biomedical visualization and imaging refers to the mechan- isms and techniques utilized to create and display images of the human body, organs or their components for clinical or research purposes. -

Point-Based Computer Graphics
Point-Based Computer Graphics Eurographics 2002 Tutorial T6 Organizers Markus Gross ETH Zürich Hanspeter Pfister MERL, Cambridge Presenters Marc Alexa TU Darmstadt Markus Gross ETH Zürich Mark Pauly ETH Zürich Hanspeter Pfister MERL, Cambridge Marc Stamminger Bauhaus-Universität Weimar Matthias Zwicker ETH Zürich Contents Tutorial Schedule................................................................................................2 Presenters Biographies........................................................................................3 Presenters Contact Information ..........................................................................4 References...........................................................................................................5 Project Pages.......................................................................................................6 Tutorial Schedule 8:30-8:45 Introduction (M. Gross) 8:45-9:45 Point Rendering (M. Zwicker) 9:45-10:00 Acquisition of Point-Sampled Geometry and Appearance I (H. Pfister) 10:00-10:30 Coffee Break 10:30-11:15 Acquisition of Point-Sampled Geometry and Appearance II (H. Pfister) 11:15-12:00 Dynamic Point Sampling (M. Stamminger) 12:00-14:00 Lunch 14:00-15:00 Point-Based Surface Representations (M. Alexa) 15:00-15:30 Spectral Processing of Point-Sampled Geometry (M. Gross) 15:30-16:00 Coffee Break 16:00-16:30 Efficient Simplification of Point-Sampled Geometry (M. Pauly) 16:30-17:15 Pointshop3D: An Interactive System for Point-Based Surface Editing (M. Pauly) 17:15-17:30 Discussion (all) 2 Presenters Biographies Dr. Markus Gross is a professor of computer science and the director of the computer graphics laboratory of the Swiss Federal Institute of Technology (ETH) in Zürich. He received a degree in electrical and computer engineering and a Ph.D. on computer graphics and image analysis, both from the University of Saarbrucken, Germany. From 1990 to 1994 Dr. Gross was with the Computer Graphics Center in Darmstadt, where he established and directed the Visual Computing Group. -

Mike Roberts Curriculum Vitae
Mike Roberts +1 (650) 924–7168 [email protected] mikeroberts3000.github.io Research Using photorealistic synthetic data for computer vision; motion planning, trajectory optimization, Interests and control methods for robotics; reconstructing 3D scenes from images; continuous and discrete optimization; submodular optimization; software tools and algorithms for creativity support Education Stanford University Stanford, California Ph.D. Computer Science 2012–2019 Advisor: Pat Hanrahan Dissertation: Trajectory Optimization Methods for Drone Cameras Harvard University Cambridge, Massachusetts Visiting Research Fellow Summer 2013 Advisor: Hanspeter Pfister University of Calgary Calgary, Canada M.S. Computer Science 2010 University of Calgary Calgary, Canada B.S. Computer Science 2007 Employment Intel Labs Seattle, Washington Research Scientist 2021– Mentor: Vladlen Koltun Apple Seattle, Washington Research Scientist 2018–2021 Microsoft Research Redmond, Washington Research Intern Summer 2016, 2017 Mentors: Neel Joshi, Sudipta Sinha Skydio Redwood City, California Research Intern Spring 2016 Mentors: Adam Bry, Frank Dellaert Udacity Mountain View, California Course Developer, Introduction to Parallel Computing 2012–2013 Instructors: John Owens, David Luebke Harvard University Cambridge, Massachusetts Research Fellow 2010–2012 Advisor: Hanspeter Pfister NVIDIA Austin, Texas Developer Tools Programmer Intern Summer 2009 Radical Entertainment Vancouver, Canada Graphics Programmer Intern 2005–2006 Honors And Featured in the Highlights of -

Mike Roberts Curriculum Vitae
Mike Roberts Union Street, Suite , Seattle, Washington, + () / [email protected] http://mikeroberts.github.io Research Using photorealistic synthetic data for computer vision; motion planning, trajectory optimization, Interests and control methods for robotics; reconstructing D scenes from images; continuous and discrete optimization; submodular optimization; software tools and algorithms for creativity support. Education Stanford University Stanford, California Ph.D. Computer Science – Advisor: Pat Hanrahan Dissertation: Trajectory Optimization Methods for Drone Cameras Harvard University Cambridge, Massachusetts Visiting Research Fellow Summer John A. Paulson School of Engineering and Applied Sciences Advisor: Hanspeter Pfister University of Calgary Calgary, Canada M.S. Computer Science University of Calgary Calgary, Canada B.S. Computer Science Employment Apple Seattle, Washington Research Scientist – Microsoft Research Redmond, Washington Research Intern Summer , Advisors: Neel Joshi, Sudipta Sinha Skydio Redwood City, California Research Intern Spring Mentors: Adam Bry, Frank Dellaert Udacity Mountain View, California Course Developer, Introduction to Parallel Computing – Instructors: John Owens, David Luebke Harvard University Cambridge, Massachusetts Research Fellow, John A. Paulson School of Engineering and Applied Sciences – Advisor: Hanspeter Pfister NVIDIA Austin, Texas Developer Tools Programmer Intern Summer Radical Entertainment Vancouver, Canada Graphics Programmer Intern – Honors And Featured in the Highlights -

Monocular Neural Image Based Rendering with Continuous View Control
Monocular Neural Image Based Rendering with Continuous View Control Xu Chen∗ Jie Song∗ Otmar Hilliges AIT Lab, ETH Zurich {xuchen, jsong, otmar.hilliges}@inf.ethz.ch ... ... Figure 1: Interactive novel view synthesis: Given a single source view our approach can generate a continuous sequence of geometrically accurate novel views under fine-grained control. Top: Given a single street-view like input, a user may specify a continuous camera trajectory and our system generates the corresponding views in real-time. Bottom: An unseen hi-res internet image is used to synthesize novel views, while the camera is controlled interactively. Please refer to our project homepagey. Abstract like to view products interactively in 3D rather than from We propose a method to produce a continuous stream discrete view angles. Likewise in map applications it is of novel views under fine-grained (e.g., 1◦ step-size) cam- desirable to explore the vicinity of street-view like images era control at interactive rates. A novel learning pipeline beyond the position at which the photograph was taken. This determines the output pixels directly from the source color. is often not possible because either only 2D imagery exists, Injecting geometric transformations, including perspective or because storing and rendering of full 3D information does projection, 3D rotation and translation into the network not scale. To overcome this limitation we study the problem forces implicit reasoning about the underlying geometry. of interactive view synthesis with 6-DoF view control, taking The latent 3D geometry representation is compact and mean- only a single image as input. We propose a method that ingful under 3D transformation, being able to produce geo- can produce a continuous stream of novel views under fine- grained (e.g., 1◦ step-size) camera control (see Fig.1). -

Visualization of Biomedical Data
Visualization of Biomedical Data Corresponding author: Seán I. O’Donoghue; email: [email protected] • Data61, Commonwealth Scientific and Industrial Research Organisation (CSIRO), Eveleigh NSW 2015, Australia • Genomics and Epigenetics Division, Garvan Institute of Medical Research, Sydney NSW 2010, Australia • School of Biotechnology and Biomolecular Sciences, UNSW, Kensington NSW 2033, Australia Benedetta Frida Baldi; email: [email protected] • Genomics and Epigenetics Division, Garvan Institute of Medical Research, Sydney NSW 2010, Australia Susan J Clark; email: [email protected] • Genomics and Epigenetics Division, Garvan Institute of Medical Research, Sydney NSW 2010, Australia Aaron E. Darling; email: [email protected] • The ithree institute, University of Technology Sydney, Ultimo NSW 2007, Australia James M. Hogan; email: [email protected] • School of Electrical Engineering and Computer Science, Queensland University of Technology, Brisbane QLD, 4000, Australia Sandeep Kaur; email: [email protected] • School of Computer Science and Engineering, UNSW, Kensington NSW 2033, Australia Lena Maier-Hein; email: [email protected] • Div. Computer Assisted Medical Interventions (CAMI), German Cancer Research Center (DKFZ), 69120 Heidelberg, Germany Davis J. McCarthy; email: [email protected] • European Molecular Biology Laboratory, European Bioinformatics Institute, Wellcome Genome Campus, CB10 1SD, Hinxton, Cambridge, UK • St. Vincent’s Institute of Medical Research, Fitzroy VIC 3065, Australia William -

Thouis R. Jones [email protected]
Thouis R. Jones [email protected] Summary Computational biologist with 20+ years' experience, including software engineering, algorithm development, image analysis, and solving challenging biomedical research problems. Research Experience Connectomics and Dense Reconstruction of Neural Connectivity Harvard University School of Engineering and Applied Sciences (2012-Present) My research is on software for automatic dense reconstruction of neural connectivity. Using using high-resolution serial sectioning and electron microscopy, biologists are able to image nanometer-scale details of neural structure. This data is extremely large (1 petabyte / cubic mm). The tools I develop in collaboration with the members of the Connectomics project allow biologists to analyze and explore this data. Image analysis, high-throughput screening, & large-scale data analysis Curie Institute, Department of Translational Research (2010 - 2012) Broad Institute of MIT and Harvard, Imaging Platform (2007 - 2010) My research focused on image and data analysis for high-content screening (HCS) and large-scale machine learning applied to HCS data. My work emphasized making powerful analysis methods available to the biological community in user-friendly tools. As part of this effort, I cofounded the open-source CellProfiler project with Dr. Anne Carpenter (director of the Imaging Platform at the Broad Institute). PhD Research: High-throughput microscopy and RNAi to reveal gene function. My doctoral research focused on methods for determining gene function using high- throughput microscopy, living cell microarrays, and RNA interference. To accomplish this goal, my collaborators and I designed and released the first open-source high- throughput cell image analysis software, CellProfiler (www.cellprofiler.org). We also developed advanced data mining methods for high-content screens. -

Human-AI Collaboration for Natural Language Generation with Interpretable Neural Networks
Human-AI Collaboration for Natural Language Generation with Interpretable Neural Networks a dissertation presented by Sebastian Gehrmann to The John A. Paulson School of Engineering and Applied Sciences in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the subject of Computer Science Harvard University Cambridge, Massachusetts May 2020 ©2019 – Sebastian Gehrmann Creative Commons Attribution License 4.0. You are free to share and adapt these materials for any purpose if you give appropriate credit and indicate changes. Thesis advisors: Barbara J. Grosz, Alexander M. Rush Sebastian Gehrmann Human-AI Collaboration for Natural Language Generation with Interpretable Neural Networks Abstract Using computers to generate natural language from information (NLG) requires approaches that plan the content and structure of the text and actualize it in fuent and error-free language. The typical approaches to NLG are data-driven, which means that they aim to solve the problem by learning from annotated data. Deep learning, a class of machine learning models based on neural networks, has become the standard data-driven NLG approach. While deep learning approaches lead to increased performance, they replicate undesired biases from the training data and make inexplicable mistakes. As a result, the outputs of deep learning NLG models cannot be trusted. Wethus need to develop ways in which humans can provide oversight over model outputs and retain their agency over an otherwise automated writing task. This dissertation argues that to retain agency over deep learning NLG models, we need to design them as team members instead of autonomous agents. We can achieve these team member models by considering the interaction design as an integral part of the machine learning model development. -

Data Visualization by Nils Gehlenborg
Data Visualization Nils Gehlenborg ([email protected]) Center for Biomedical Informatics / Harvard Medical School Cancer Program / Broad Institute of MIT and Harvard ISMB/ECCB 2011 http://www.biovis.net Flyers at ISCB booth! Data Visualization / ISMB/ECCB 2011 / Nils Gehlenborg A good sketch is better than a long speech. Napoleon Bonaparte Data Visualization / ISMB/ECCB 2011 / Nils Gehlenborg Minard 1869 Napoleon’s March on Moscow Data Visualization / ISMB/ECCB 2011 / Nils Gehlenborg 4 I believe when I see it. Unknown Data Visualization / ISMB/ECCB 2011 / Nils Gehlenborg Anscombe 1973, The American Statistician Anscombe’s Quartet mean(X) = 9, var(X) = 11, mean(Y) = 7.5, var(Y) = 4.12, cor(X,Y) = 0.816, linear regression line Y = 3 + 0.5*X Data Visualization / ISMB/ECCB 2011 / Nils Gehlenborg 6 Anscombe 1973, The American Statistician Anscombe’s Quartet Data Visualization / ISMB/ECCB 2011 / Nils Gehlenborg 7 Exploration: Hypothesis Generation trends gaps outliers clusters - A large data set is given and the goal is to learn something about it. - Visualization is employed to perform pattern detection using the human visual system. - The goal is to generate hypotheses that can be tested with statistical methods or follow-up experiments. Data Visualization / ISMB/ECCB 2011 / Nils Gehlenborg 8 Visualization Use Cases Presentation Confirmation Exploration Data Visualization / ISMB/ECCB 2011 / Nils Gehlenborg 9 Definition The use of computer-supported, interactive, visual representations of data to amplify cognition. Stu Card, Jock Mackinlay & Ben Shneiderman Computer-based visualization systems provide visual representations of datasets intended to help people carry out some task more effectively.effectively. -

Pop-Out, Illusions, Interaction
Pop-Out, Illusions, Interaction DS 4200 FALL 2020 Prof. Cody Dunne NORTHEASTERN UNIVERSITY Slides and inspiration from Michelle Borkin, Krzysztof Gajos, Hanspeter Pfister, Miriah Meyer, Jonathan Schwabish, and David Sprague 1 CHECK-IN 2 PREVIOUSLY, ON DS 4200… 4 Color Vocabulary and Perceptual Ordering Darkness (Lightness) Saturation Hue ? ? Based on Slides by Miriah Meyer, Tamara Munzner 5 Color Deficiencies (Color Blindness) Based on Slides by Hanspeter Pfister, Maureen Stone 6 Check your images/colormaps for issues! http://www.vischeck.com/vischeck/vischeckImage.php https://www.color-blindness.com/coblis-color-blindness-simulator/ 7 Color Brewer http://colorbrewer2.org 8 Color Advice Summary Use a limited hue palette • Control color “pop out” with low-saturation colors • Avoid clutter from too many competing colors Use neutral backgrounds • Control impact of color • Minimize simultaneous contrast Use Color Brewer etc. for picking scales Don’t forget aesthetics! Based on Slides by Hanspeter Pfister, Maureen Stone 9 NOW, ON DS 4200… 10 POP-OUT EFFECTS 15 POP-OUT EFFECTS COLOR Healey, 2012 16 POP-OUT EFFECTS SHAPE Healey, 2012 17 POP-OUT EFFECTS “CONJUNCTION” (HARDER TO FIND RED CIRCLE!) Healey, 2012 18 POP-OUT EFFECTS MOTION Healey, 2012 19 POP-OUT EFFECTS Healey, 2012 20 POP-OUT EFFECTS Healey, 2012 21 Use these “popout” effects to help design effective visualizations! (E.g., draw viewer’s attention to main points, effective redundant encodings, etc.) Ware, VTFD 22 Discriminability and Separability The question of discriminability is: if you encode data using a particular visual channel, are the differences between items perceptible to the human as intended? Munzner, VAD 23 Textures easy hard Ware, VTFD 25 Textures: Interference Ware, VTFD 26 ILLUSIONS AND TRICKS 28 Visual Attention & Change Blindness 30 Visual Attention & Change Blindness Task: Identify the lumps/nodules in the patient’s lungs to look for cancer or abnormal growth.