Tissue Signals Imprint ILC2 Identity with Anticipatory Function
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

IL17RB Antibody Cat
IL17RB Antibody Cat. No.: 62-448 IL17RB Antibody Formalin-fixed and paraffin-embedded human colon Flow cytometric analysis of HepG2 cells using IL17RB carcinoma reacted with IL17RB Antibody , which was Antibody (bottom histogram) compared to a negative peroxidase-conjugated to the secondary antibody, control cell (top histogram). FITC-conjugated goat-anti- followed by DAB staining. rabbit secondary antibodies were used for the analysis. Specifications HOST SPECIES: Rabbit SPECIES REACTIVITY: Human This IL17_ antibody is generated from rabbits immunized with a KLH conjugated synthetic IMMUNOGEN: peptide between 207-234 amino acids from the Central region of human IL17_. TESTED APPLICATIONS: Flow, IHC-P, WB For WB starting dilution is: 1:1000 APPLICATIONS: For IHC-P starting dilution is: 1:10~50 For FACS starting dilution is: 1:10~50 September 25, 2021 1 https://www.prosci-inc.com/il17rb-antibody-62-448.html PREDICTED MOLECULAR 56 kDa WEIGHT: Properties This antibody is prepared by Saturated Ammonium Sulfate (SAS) precipitation followed by PURIFICATION: dialysis CLONALITY: Polyclonal ISOTYPE: Rabbit Ig CONJUGATE: Unconjugated PHYSICAL STATE: Liquid BUFFER: Supplied in PBS with 0.09% (W/V) sodium azide. CONCENTRATION: batch dependent Store at 4˚C for three months and -20˚C, stable for up to one year. As with all antibodies STORAGE CONDITIONS: care should be taken to avoid repeated freeze thaw cycles. Antibodies should not be exposed to prolonged high temperatures. Additional Info OFFICIAL SYMBOL: IL17RB Interleukin-17 receptor B, IL-17 receptor B, IL-17RB, Cytokine receptor-like 4, IL-17 ALTERNATE NAMES: receptor homolog 1, IL-17Rh1, IL17Rh1, Interleukin-17B receptor, IL-17B receptor, IL17RB, CRL4, EVI27, IL17BR ACCESSION NO.: Q9NRM6 PROTEIN GI NO.: 21263748 GENE ID: 55540 USER NOTE: Optimal dilutions for each application to be determined by the researcher. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Table S1| Differential Expression Analysis of the Atopy Transcriptome
Table S1| Differential expression analysis of the atopy transcriptome in CD4+ T-cell responses to allergens in atopic and nonatopic subjects Probe ID S.test Gene Symbol Gene Description Chromosome Statistic Location 7994280 10.32 IL4R Interleukin 4 receptor 16p11.2-12.1 8143383 8.95 --- --- --- 7974689 8.50 DACT1 Dapper, antagonist of beta-catenin, homolog 1 14q23.1 8102415 7.59 CAMK2D Calcium/calmodulin-dependent protein kinase II delta 4q26 7950743 7.58 RAB30 RAB30, member RAS oncogene family 11q12-q14 8136580 7.54 RAB19B GTP-binding protein RAB19B 7q34 8043504 7.45 MAL Mal, T-cell differentiation protein 2cen-q13 8087739 7.27 CISH Cytokine inducible SH2-containing protein 3p21.3 8000413 7.17 NSMCE1 Non-SMC element 1 homolog (S. cerevisiae) 16p12.1 8021301 7.15 RAB27B RAB27B, member RAS oncogene family 18q21.2 8143367 6.83 SLC37A3 Solute carrier family 37 member 3 7q34 8152976 6.65 TMEM71 Transmembrane protein 71 8q24.22 7931914 6.56 IL2R Interleukin 2 receptor, alpha 10p15-p14 8014768 6.43 PLXDC1 Plexin domain containing 1 17q21.1 8056222 6.43 DPP4 Dipeptidyl-peptidase 4 (CD26) 2q24.3 7917697 6.40 GFI1 Growth factor independent 1 1p22 7903507 6.39 FAM102B Family with sequence similarity 102, member B 1p13.3 7968236 5.96 RASL11A RAS-like, family 11, member A --- 7912537 5.95 DHRS3 Dehydrogenase/reductase (SDR family) member 3 1p36.1 7963491 5.83 KRT1 Keratin 1 (epidermolytic hyperkeratosis) 12q12-q13 7903786 5.72 CSF1 Colony stimulating factor 1 (macrophage) 1p21-p13 8019061 5.67 SGSH N-sulfoglucosamine sulfohydrolase (sulfamidase) 17q25.3 -

Table SI. Characteristics of 9 Patients Chosen from 60 Patients with Hepatocellular Carcinoma
Table SI. Characteristics of 9 patients chosen from 60 patients with hepatocellular carcinoma. Characteristic No. of patients (%) Age, years <50 3 (33.3) ≥50 6 (66.7) Sex Female 4 (44.4) Male 5 (55.6) Albumin, g/l ≥35 8 (88.9) <35 1 (11.1) AFP, ng/ml ≤20 2 (22.2) >20 7 (77.8) Tumor diameter, cm ≤5 3 (33.3) >5 6 (66.7) Portal vein invasion Without 3 (33.3) With 6 (66.7) BCLC stage A 2 (22.2) B/C 7 (77.8) AFP, α-fetoprotein; BCLC, Barcelona Clinic Liver Cancer. Table SII. List of the gene symbol and relative expression of six significant clusters. Gene symbol SPOT Profile Day 0 Day 3 Day 7 Day 14 Day 21 ALCAM ID_10 16 0 -356.5 378.5 0.1 307.8 ANGPT2 ID_13 16 0 -45.7 197.2 96 60.2 CD80 ID_24 16 0 -177 362 -29 187 CTNNB1 ID_29 16 0 -58 294 83 49 BMP4 ID_37 16 0 -107 189 0 53 CCL28 ID_49 16 0 -307 856 491 347 CCR1 ID_50 16 0 -189 391 236 112 CCR4 ID_53 16 0 -101 361 89 240 CCR7 ID_56 16 0 -98 361 103 66 CLC ID_70 16 0 -147 274 0 188 CRIM1 ID_74 16 0 -642 229 -48 -112 CXCL14 ID_83 16 0 -434 1,154.00 778 376 CXCL16 ID_84 16 0 -128 800 477 180 DKK3 ID_97 16 0 -199 610 107 3 FGFBP1 ID_129 16 0 -224 484 -155 37 FGF20 ID_147 16 0 -113 264 -32 -4 WFIKKN1 ID_163 16 0 -195 544 262 41 GFRA1 ID_175 16 0 -536 443 29 280 GFRA4 ID_178 16 0 -211 208 -47 116 TNFSF18 ID_180 16 0 -14 246 89 68 GPC5 ID_187 16 0 -180 1,345.00 298 509 GH1 ID_194 16 0 -596 206 10 -9 CCHCR1 ID_198 16 0 -108 322 50 238 NRG1 ID_200 16 0 -81 378.5 170.5 187 ICAM1 ID_207 16 0 -136 98 -17 63 IFNB1 ID_213 16 0 -87 233 143 29 IGFBP3 ID_218 16 0 -1,311.00 609 -368 -178 IGFBP4 ID_219 16 0 -

Supplementary Material DNA Methylation in Inflammatory Pathways Modifies the Association Between BMI and Adult-Onset Non- Atopic
Supplementary Material DNA Methylation in Inflammatory Pathways Modifies the Association between BMI and Adult-Onset Non- Atopic Asthma Ayoung Jeong 1,2, Medea Imboden 1,2, Akram Ghantous 3, Alexei Novoloaca 3, Anne-Elie Carsin 4,5,6, Manolis Kogevinas 4,5,6, Christian Schindler 1,2, Gianfranco Lovison 7, Zdenko Herceg 3, Cyrille Cuenin 3, Roel Vermeulen 8, Deborah Jarvis 9, André F. S. Amaral 9, Florian Kronenberg 10, Paolo Vineis 11,12 and Nicole Probst-Hensch 1,2,* 1 Swiss Tropical and Public Health Institute, 4051 Basel, Switzerland; [email protected] (A.J.); [email protected] (M.I.); [email protected] (C.S.) 2 Department of Public Health, University of Basel, 4001 Basel, Switzerland 3 International Agency for Research on Cancer, 69372 Lyon, France; [email protected] (A.G.); [email protected] (A.N.); [email protected] (Z.H.); [email protected] (C.C.) 4 ISGlobal, Barcelona Institute for Global Health, 08003 Barcelona, Spain; [email protected] (A.-E.C.); [email protected] (M.K.) 5 Universitat Pompeu Fabra (UPF), 08002 Barcelona, Spain 6 CIBER Epidemiología y Salud Pública (CIBERESP), 08005 Barcelona, Spain 7 Department of Economics, Business and Statistics, University of Palermo, 90128 Palermo, Italy; [email protected] 8 Environmental Epidemiology Division, Utrecht University, Institute for Risk Assessment Sciences, 3584CM Utrecht, Netherlands; [email protected] 9 Population Health and Occupational Disease, National Heart and Lung Institute, Imperial College, SW3 6LR London, UK; [email protected] (D.J.); [email protected] (A.F.S.A.) 10 Division of Genetic Epidemiology, Medical University of Innsbruck, 6020 Innsbruck, Austria; [email protected] 11 MRC-PHE Centre for Environment and Health, School of Public Health, Imperial College London, W2 1PG London, UK; [email protected] 12 Italian Institute for Genomic Medicine (IIGM), 10126 Turin, Italy * Correspondence: [email protected]; Tel.: +41-61-284-8378 Int. -

Single-Cell Analysis of Crohn's Disease Lesions Identifies
bioRxiv preprint doi: https://doi.org/10.1101/503102; this version posted December 20, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy JC Martin1,2,3, G Boschetti1,2,3, C Chang1,2,3, R Ungaro4, M Giri5, LS Chuang5, S Nayar5, A Greenstein6, M. Dubinsky7, L Walker1,2,5,8, A Leader1,2,3, JS Fine9, CE Whitehurst9, L Mbow9, S Kugathasan10, L.A. Denson11, J.Hyams12, JR Friedman13, P Desai13, HM Ko14, I Laface1,2,8, Guray Akturk1,2,8, EE Schadt15,16, S Gnjatic1,2,8, A Rahman1,2,5,8, , M Merad1,2,3,8,17,18*, JH Cho5,17,*, E Kenigsberg1,15,16,17* 1 Precision Immunology Institute, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA. 2 Tisch Cancer Institute, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA. 3 Department of Oncological Sciences, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA. 4 The Dr. Henry D. Janowitz Division of Gastroenterology, Icahn School of Medicine at Mount Sinai, New York City, NY 10029, USA. 5 Charles Bronfman Institute for Personalized Medicine, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA. 6 Department of Colorectal Surgery, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA 7 Department of Pediatrics, Susan and Leonard Feinstein IBD Clinical Center, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA. -

Evolutionary Divergence and Functions of the Human Interleukin (IL) Gene Family Chad Brocker,1 David Thompson,2 Akiko Matsumoto,1 Daniel W
UPDATE ON GENE COMPLETIONS AND ANNOTATIONS Evolutionary divergence and functions of the human interleukin (IL) gene family Chad Brocker,1 David Thompson,2 Akiko Matsumoto,1 Daniel W. Nebert3* and Vasilis Vasiliou1 1Molecular Toxicology and Environmental Health Sciences Program, Department of Pharmaceutical Sciences, University of Colorado Denver, Aurora, CO 80045, USA 2Department of Clinical Pharmacy, University of Colorado Denver, Aurora, CO 80045, USA 3Department of Environmental Health and Center for Environmental Genetics (CEG), University of Cincinnati Medical Center, Cincinnati, OH 45267–0056, USA *Correspondence to: Tel: þ1 513 821 4664; Fax: þ1 513 558 0925; E-mail: [email protected]; [email protected] Date received (in revised form): 22nd September 2010 Abstract Cytokines play a very important role in nearly all aspects of inflammation and immunity. The term ‘interleukin’ (IL) has been used to describe a group of cytokines with complex immunomodulatory functions — including cell proliferation, maturation, migration and adhesion. These cytokines also play an important role in immune cell differentiation and activation. Determining the exact function of a particular cytokine is complicated by the influence of the producing cell type, the responding cell type and the phase of the immune response. ILs can also have pro- and anti-inflammatory effects, further complicating their characterisation. These molecules are under constant pressure to evolve due to continual competition between the host’s immune system and infecting organisms; as such, ILs have undergone significant evolution. This has resulted in little amino acid conservation between orthologous proteins, which further complicates the gene family organisation. Within the literature there are a number of overlapping nomenclature and classification systems derived from biological function, receptor-binding properties and originating cell type. -

The Innate Cytokines IL-25, IL-33, and TSLP Cooperate in the Induction Of
The Innate Cytokines IL-25, IL-33, and TSLP Cooperate in the Induction of Type 2 Innate Lymphoid Cell Expansion and Mucous Metaplasia in Rhinovirus-Infected This information is current as Immature Mice of September 29, 2021. Mingyuan Han, Charu Rajput, Jun Y. Hong, Jing Lei, Joanna L. Hinde, Qian Wu, J. Kelley Bentley and Marc B. Hershenson J Immunol 2017; 199:1308-1318; Prepublished online 12 Downloaded from July 2017; doi: 10.4049/jimmunol.1700216 http://www.jimmunol.org/content/199/4/1308 http://www.jimmunol.org/ Supplementary http://www.jimmunol.org/content/suppl/2017/07/12/jimmunol.170021 Material 6.DCSupplemental References This article cites 64 articles, 15 of which you can access for free at: http://www.jimmunol.org/content/199/4/1308.full#ref-list-1 by guest on September 29, 2021 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2017 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. -

KRAS Mutations Are Negatively Correlated with Immunity in Colon Cancer
www.aging-us.com AGING 2021, Vol. 13, No. 1 Research Paper KRAS mutations are negatively correlated with immunity in colon cancer Xiaorui Fu1,2,*, Xinyi Wang1,2,*, Jinzhong Duanmu1, Taiyuan Li1, Qunguang Jiang1 1Department of Gastrointestinal Surgery, The First Affiliated Hospital of Nanchang University, Nanchang, Jiangxi, People's Republic of China 2Queen Mary College, Medical Department, Nanchang University, Nanchang, Jiangxi, People's Republic of China *Equal contribution Correspondence to: Qunguang Jiang; email: [email protected] Keywords: KRAS mutations, immunity, colon cancer, tumor-infiltrating immune cells, inflammation Received: March 27, 2020 Accepted: October 8, 2020 Published: November 26, 2020 Copyright: © 2020 Fu et al. This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT The heterogeneity of colon cancer tumors suggests that therapeutics targeting specific molecules may be effective in only a few patients. It is therefore necessary to explore gene mutations in colon cancer. In this study, we obtained colon cancer samples from The Cancer Genome Atlas, and the International Cancer Genome Consortium. We evaluated the landscape of somatic mutations in colon cancer and found that KRAS mutations, particularly rs121913529, were frequent and had prognostic value. Using ESTIMATE analysis, we observed that the KRAS-mutated group had higher tumor purity, lower immune score, and lower stromal score than the wild- type group. Through single-sample Gene Set Enrichment Analysis and Gene Set Enrichment Analysis, we found that KRAS mutations negatively correlated with enrichment levels of tumor infiltrating lymphocytes, inflammation, and cytolytic activities. -

Lineage Tracing and Targeting of IL17RB+ Tuft Cell-Like Human
+ Lineage tracing and targeting of IL17RB tuft cell-like human colorectal cancer stem cells Norihiro Gotoa, Akihisa Fukudaa, Yuichi Yamagaa, Takaaki Yoshikawaa, Takahisa Marunoa, Hisatsugu Maekawab,c, Susumu Inamotob, Kenji Kawadab, Yoshiharu Sakaib, Hiroyuki Miyoshic, Makoto Mark Taketoc, Tsutomu Chibad, and Hiroshi Senoa,1 aDepartment of Gastroenterology and Hepatology, Kyoto University Graduate School of Medicine, 606-8507 Kyoto, Japan; bDepartment of Surgery, Kyoto University Graduate School of Medicine, 606-8507 Kyoto, Japan; cDivision of Experimental Therapeutics, Kyoto University Graduate School of Medicine, 606-8507 Kyoto, Japan; and dDepartment of Gastroenterology and Hepatology, Kansai Electric Power Hospital, 553-0003 Osaka, Japan Edited by Napoleone Ferrara, University of California San Diego, La Jolla, CA, and approved May 16, 2019 (received for review January 7, 2019) Cancer stem cell (CSC)-specific markers may be potential thera- Recent studies showed that the tuft cell is the main source of IL- peutic targets. We previously identified that Dclk1, a tuft cell 25 in the intestine (13–15) and its receptor IL17RB is expressed marker, marks tumor stem cells (TSCs) in mouse intestinal adeno- by type 2 innate lymphoid cells (ILC2s) in the lamina propria + mas. Based on the analysis of mouse Dclk1 tumor cells, we aimed (16). In case of helminth infection, induction of ILC2s by tuft to identify a CSC-specific cell surface marker in human colorectal cell-derived IL-25 results in IL-13 production and subsequent cancers (hCRCs) and validate the therapeutic effect of targeting it. + tuft cell and goblet cell hyperplasia for worm expulsion (13, 15). IL17RB was distinctively expressed by Dclk1 mouse intestinal tu- Although it has been indicated that IL17RB is also expressed in Il17rb-CreERT2-IRES-EGFP mor cells. -
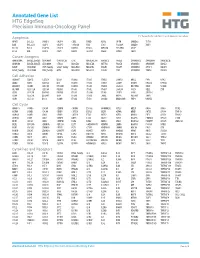
Annotated Gene List HTG Edgeseq Precision Immuno-Oncology Panel
Annotated Gene List HTG EdgeSeq Precision Immuno-Oncology Panel For Research Use Only. Not for use in diagnostic procedures. Apoptosis APAF1 BCL2L1 CARD11 CASP4 CD5L FADD KSR2 OPTN SAMD12 TCF19 BAX BCL2L11 CASP1 CASP5 CORO1A FAS LRG1 PLA2G6 SAMD9 XAF1 BCL10 BCL6 CASP10 CASP8 DAPK2 FASLG MECOM PYCARD SPOP BCL2 BID CASP3 CAV1 DAPL1 GLIPR1 MELK RIPK2 TBK1 Cancer Antigens ANKRD30A BAGE2_BAGE3 CEACAM6 CTAG1A_1B LIPE MAGEA3_A6 MAGEC2 PAGE3 SPANXACD SPANXN4 XAGE1B_1E ARMCX6 BAGE4_BAGE5 CEACAM8 CTAG2 MAGEA1 MAGEA4 MTFR2 PAGE4 SPANXB1 SPANXN5 XAGE2 BAGE CEACAM1 CT45_family GAGE_family MAGEA10 MAGEB2 PAGE1 PAGE5 SPANXN1 SYCP1 XAGE3 BAGE_family CEACAM5 CT47_family HPN MAGEA12 MAGEC1 PAGE2 PBK SPANXN3 TEX14 XAGE5 Cell Adhesion ADAM17 CDH15 CLEC5A DSG3 ICAM2 ITGA5 ITGB2 LAMC3 MBL2 PVR UPK2 ADD2 CDH5 CLEC6A DST ICAM3 ITGA6 ITGB3 LAMP1 MTDH RRAS2 UPK3A ADGRE5 CLDN3 CLEC7A EPCAM ICAM4 ITGAE ITGB4 LGALS1 NECTIN2 SELE VCAM1 ALCAM CLEC12A CLEC9A FBLN1 ITGA1 ITGAL ITGB7 LGALS3 OCLN SELL ZYX CD63 CLEC2B DIAPH3 FXYD5 ITGA2 ITGAM ITLN2 LYVE1 OLR1 SELPLG CD99 CLEC4A DLGAP5 IBSP ITGA3 ITGAX JAML M6PR PECAM1 THY1 CDH1 CLEC4C DSC3 ICAM1 ITGA4 ITGB1 L1CAM MADCAM1 PKP1 UNC5D Cell Cycle ANAPC1 CCND3 CDCA5 CENPH CNNM1 ESCO2 HORMAD2 KIF2C MELK ORC6 SKA3 TPX2 ASPM CCNE1 CDCA8 CENPI CNTLN ESPL1 IKZF1 KIF4A MND1 PATZ1 SP100 TRIP13 AURKA CCNE2 CDK1 CENPL CNTLN ETS1 IKZF2 KIF5C MYBL2 PIF1 SP110 TROAP AURKB CCNF CDK4 CENPU DBF4 ETS2 IKZF3 KIFC1 NCAPG PIMREG SPC24 TUBB BEX1 CDC20 CDK6 CENPW E2F2 EZH2 IKZF4 KNL1 NCAPG2 PKMYT1 SPC25 ZWILCH BEX2 CDC25A CDKN1A CEP250 E2F7 GADD45GIP1 -

Xo PANEL DNA GENE LIST
xO PANEL DNA GENE LIST ~1700 gene comprehensive cancer panel enriched for clinically actionable genes with additional biologically relevant genes (at 400 -500x average coverage on tumor) Genes A-C Genes D-F Genes G-I Genes J-L AATK ATAD2B BTG1 CDH7 CREM DACH1 EPHA1 FES G6PC3 HGF IL18RAP JADE1 LMO1 ABCA1 ATF1 BTG2 CDK1 CRHR1 DACH2 EPHA2 FEV G6PD HIF1A IL1R1 JAK1 LMO2 ABCB1 ATM BTG3 CDK10 CRK DAXX EPHA3 FGF1 GAB1 HIF1AN IL1R2 JAK2 LMO7 ABCB11 ATR BTK CDK11A CRKL DBH EPHA4 FGF10 GAB2 HIST1H1E IL1RAP JAK3 LMTK2 ABCB4 ATRX BTRC CDK11B CRLF2 DCC EPHA5 FGF11 GABPA HIST1H3B IL20RA JARID2 LMTK3 ABCC1 AURKA BUB1 CDK12 CRTC1 DCUN1D1 EPHA6 FGF12 GALNT12 HIST1H4E IL20RB JAZF1 LPHN2 ABCC2 AURKB BUB1B CDK13 CRTC2 DCUN1D2 EPHA7 FGF13 GATA1 HLA-A IL21R JMJD1C LPHN3 ABCG1 AURKC BUB3 CDK14 CRTC3 DDB2 EPHA8 FGF14 GATA2 HLA-B IL22RA1 JMJD4 LPP ABCG2 AXIN1 C11orf30 CDK15 CSF1 DDIT3 EPHB1 FGF16 GATA3 HLF IL22RA2 JMJD6 LRP1B ABI1 AXIN2 CACNA1C CDK16 CSF1R DDR1 EPHB2 FGF17 GATA5 HLTF IL23R JMJD7 LRP5 ABL1 AXL CACNA1S CDK17 CSF2RA DDR2 EPHB3 FGF18 GATA6 HMGA1 IL2RA JMJD8 LRP6 ABL2 B2M CACNB2 CDK18 CSF2RB DDX3X EPHB4 FGF19 GDNF HMGA2 IL2RB JUN LRRK2 ACE BABAM1 CADM2 CDK19 CSF3R DDX5 EPHB6 FGF2 GFI1 HMGCR IL2RG JUNB LSM1 ACSL6 BACH1 CALR CDK2 CSK DDX6 EPOR FGF20 GFI1B HNF1A IL3 JUND LTK ACTA2 BACH2 CAMTA1 CDK20 CSNK1D DEK ERBB2 FGF21 GFRA4 HNF1B IL3RA JUP LYL1 ACTC1 BAG4 CAPRIN2 CDK3 CSNK1E DHFR ERBB3 FGF22 GGCX HNRNPA3 IL4R KAT2A LYN ACVR1 BAI3 CARD10 CDK4 CTCF DHH ERBB4 FGF23 GHR HOXA10 IL5RA KAT2B LZTR1 ACVR1B BAP1 CARD11 CDK5 CTCFL DIAPH1 ERCC1 FGF3 GID4 HOXA11