The Population Structure of Ten Newfoundland Outports
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Community Needs and Resources Assessment for the Port Aux Basques and Burgeo Areas
PRIMARY HEALTH CARE IN ACTION A Community Needs and Resources Assessment for the Port aux Basques and Burgeo Areas 2013 Prepared by: Danielle Shea, RD, M.Ad.Ed. Primary Health Care Manager, Bay St. George Area Table of Contents Executive Summary Page 4 Community Health Needs and Resources Assessment Page 6 Survey Overview Page 6 Survey Results Page 7 Demographics Page 7 Community Services Page 8 Health Related Community Services Page 10 Community Groups Page 15 Community Concerns Page 16 Other Page 20 Focus Group Overview Page 20 Port aux Basques: Cancer Care Page 21 Highlights Page 22 Burgeo: Healthy Eating Page 23 Highlights Page 24 Port aux Basques and Burgeo Areas Overview Page 26 Statistical Data Overview Page 28 Statistical Data Page 28 Community Resource Listing Overview Page 38 Port aux Basques Community Resource Listing Page 38 Burgeo Community Resource Listing Page 44 Strengths Page 50 Recommendations Page 51 Conclusion Page 52 References Page 54 Appendix A Page 55 Primary Health Care Model Appendix B Page 57 Community Health Needs and Resources Assessment Policy Community Health Needs and Resources Assessment Port aux Basques/ Burgeo Area Page 2 Appendix C Page 62 Community Health Needs and Resources Assessment Survey Appendix D Page 70 Port aux Basques Focus Group Questions Appendix E Page 72 Burgeo Focus Group Questions Community Health Needs and Resources Assessment Port aux Basques/ Burgeo Area Page 3 Executive Summary Primary health care is defined as an individual’s first contact with the health system and includes the full range of services from health promotion, diagnosis, and treatment to chronic disease management. -

Acknowledgment
Acknowledgment I would like to thank the following people for their help and support in the production of this book: National Literacy Secretariat for giving us funding. NLRDC for sending in the proposal. Ed Oldford and Diane Hunt of the Central - Eastern Literacy Outreach Office for their help and encouragement. Terry Morrison for his advice and help. Tony Collins and Marina John for proof reading, and Janet Power for being there. Roy Powell for drawing the sketches. The Gander Lion's Club for donating the space for our workshop. Cable 9 for donating the use of their video equipment. Rob Brown and Dean Layte for operating the video equipment. Finally and most important, the Adult Learners who contributed stories, poems, recipes and pictures: Amanda White Barbara Collins Barry Vineham Caville Tarrant Cecil Godwin Elaine Woodford John Philpott Kathleen Ford Marina Starkes Mary Mouland Maxine Steel Noreen & Rex Culter Robert Tulk Roy Powell Table of Contents Part One Personal Stories Amanda White Barbara Collins Barry Vineham Caville Tarrant Cecil Godwin Elaine Woodford John Philpott Kathleen Ford Marina Starkes Mary Mouland Maxine Steel Noreen & Rex Culter Robert Tulk Roy Powell Part Two Stories from our past Eric Hancock's Seal Hunt Barbara Collins Stuck in the Ice Barry Vineham To the Fogo I Left Behind Caville Tarrant In the Army Barry Vineham The Fifty Dollar House Marina Starkes Baber's Light Barry Vineham My Growing Up Days Maxine Steel Squid Barry Vineham A Trip to the Well Barry Vineham Two Seagulls Caville Tarrant Around the Home -

The Fogo Island Cooperative Society and Its Workers: Lessons from the Fishery”
The Fogo Island Cooperative Society and its Workers: Lessons from the Fishery” by Judy and Larry Haiven, Saint Mary's University for the Symposium “The Co-operative and its Workers” June 15-17, 2006 Introduction This story plays itself out against the background of an industry and a province torn by economic crisis. Fogo is the largest of the smaller islands of Newfoundland, found off the northeast coast. It is approximately 254 sq km (approximately 15 x 15 km if the island were an exact square). The current population is 2800; the population before the collapse of the cod fishery in 1992 was almost twice that. The story of the Fogo Island Cooperative Society began in 1967 while the cod fishery was still viable, but first a bit of history just for context. Tradition from the 15th to the 19th century was that small groups of fisher people (English and Irish with a smattering of Portuguese, Basque and French) would set up a village in a friendly cove where they would fish the sea (primarily of the more-than-plentiful cod) and live very independently. Most of the fishing was close to shore, hence the “inshore fishery.” When the cove and the land surrounding it proved too small to provide for the growing community, a group would pick up and move to another location. Thus emerged a series of small fishing villages strung out along the rocky coastline, which, depending on how closely you track all of the inlets and headlands, is as “short” as 9,000 km or as long as 17,000 km. -

House of Assembly
HOUSE OF ASSEMBLY SECOND SESSION THIRTY-SEVENTH GENERAL ASSEMBLY OF NEWFOUNDLAND 1977 >l, .: V Published under the authority of the Speaker of the House of Assembly Honourable Gerald Ryan Ottenheimer Detailed Index of Verbatim Report 2nd Session Thirty-Seventh General Assembly of Newfound I and From Fehrur” 2n’,l°77 o ?Tovether 24th, 1°77 Compiled by: Sara J. MacGillivray Hansard Division House of Assembly TABLE OF CONTENTS PAGES OFFICIAL OPENING 37th GENERAL ASSEMBLY OF NEWFOUNDLAND (2nd Session) 1—2 ADDRESS IN REPLY AND AMENDMENTS thereto 3—7 ANSWERS TO QUESTIONS 8-27 BILLS 28—45 BUDGET DEBATE 46 COMMITTEE OF SUPPLY (Estimates) 47-66 MISCELLANEOUS 67-70 ORAL QUESTIONS 71-132 PETITIONS 133—142 REPORTS, REGULATIONS, etc. 143—147 RESOLUTIONS 148—160 RULINGS 161—180 STATEMENTS (Ministerial) etc. 181—186 OFFICIAL CLOSING 187 OFFICIAL OPENING (PAGES 1—2) OFFICIAL OPENING - THIRTY-SEVENTH GENERAL ASSEMBLY OF NEWFOuNDLAND (SECOND SESSION) DATE - February 2, 1977 PAGES BOOK THRONE SPEECH - Read by it. Gov. G.A. Winter 1—14. 1. MOVER - C. Cross, Y.H.A. Bonavista North 15—19. “ SECONDER - H. Twomey, M.H.A. Exploits 20-23. “ H.M. LOYAL OPPOSITION LEADER - Hon. E.M. Roberts 23-45. Congratulations to Newfoundland’s first woman Rhodes Scholar (Miss Jacqueline Sheppard) 24. PREMIER OF NEWFOUNDLAND — Hon. F.D. Moores 45—64. MOTION:- “Select Committee be appointed to draft Address in Reply’:— C. Cross, M.H.A. Bonavista North H. Twomey, M.B.A. Exploits M. O’Brien, M.B.A. Ferryland 64. NOTICE OF RESOLUTIONS: (I. Strachan) ‘Select Committee to study and improve the role played by Labrador people” 66. -

The Hitch-Hiker Is Intended to Provide Information Which Beginning Adult Readers Can Read and Understand
CONTENTS: Foreword Acknowledgements Chapter 1: The Southwestern Corner Chapter 2: The Great Northern Peninsula Chapter 3: Labrador Chapter 4: Deer Lake to Bishop's Falls Chapter 5: Botwood to Twillingate Chapter 6: Glenwood to Gambo Chapter 7: Glovertown to Bonavista Chapter 8: The South Coast Chapter 9: Goobies to Cape St. Mary's to Whitbourne Chapter 10: Trinity-Conception Chapter 11: St. John's and the Eastern Avalon FOREWORD This book was written to give students a closer look at Newfoundland and Labrador. Learning about our own part of the earth can help us get a better understanding of the world at large. Much of the information now available about our province is aimed at young readers and people with at least a high school education. The Hitch-Hiker is intended to provide information which beginning adult readers can read and understand. This work has a special feature we hope readers will appreciate and enjoy. Many of the places written about in this book are seen through the eyes of an adult learner and other fictional characters. These characters were created to help add a touch of reality to the printed page. We hope the characters and the things they learn and talk about also give the reader a better understanding of our province. Above all, we hope this book challenges your curiosity and encourages you to search for more information about our land. Don McDonald Director of Programs and Services Newfoundland and Labrador Literacy Development Council ACKNOWLEDGMENTS I wish to thank the many people who so kindly and eagerly helped me during the production of this book. -

(PL-557) for NPA 879 to Overlay NPA
Number: PL- 557 Date: 20 January 2021 From: Canadian Numbering Administrator (CNA) Subject: NPA 879 to Overlay NPA 709 (Newfoundland & Labrador, Canada) Related Previous Planning Letters: PL-503, PL-514, PL-521 _____________________________________________________________________ This Planning Letter supersedes all previous Planning Letters related to NPA Relief Planning for NPA 709 (Newfoundland and Labrador, Canada). In Telecom Decision CRTC 2021-13, dated 18 January 2021, Indefinite deferral of relief for area code 709 in Newfoundland and Labrador, the Canadian Radio-television and Telecommunications Commission (CRTC) approved an NPA 709 Relief Planning Committee’s report which recommended the indefinite deferral of implementation of overlay area code 879 to provide relief to area code 709 until it re-enters the relief planning window. Accordingly, the relief date of 20 May 2022, which was identified in Planning Letter 521, has been postponed indefinitely. The relief method (Distributed Overlay) and new area code 879 will be implemented when relief is required. Background Information: In Telecom Decision CRTC 2017-35, dated 2 February 2017, the Canadian Radio-television and Telecommunications Commission (CRTC) directed that relief for Newfoundland and Labrador area code 709 be provided through a Distributed Overlay using new area code 879. The new area code 879 has been assigned by the North American Numbering Plan Administrator (NANPA) and will be implemented as a Distributed Overlay over the geographic area of the province of Newfoundland and Labrador currently served by the 709 area code. The area code 709 consists of 211 Exchange Areas serving the province of Newfoundland and Labrador which includes the major communities of Corner Brook, Gander, Grand Falls, Happy Valley – Goose Bay, Labrador City – Wabush, Marystown and St. -

Geology of the Connaigre Peninsula and Adjacent
10′ 55° 00′ LEGEND 32 MIDDLE PALEOZOIC LATE NEOPROTEROZOIC 42 42 DEVONIAN LONG HARBOUR GROUP (Units 16 to 24) 86 Mo BELLEORAM GRANITE Rencontre Formation (Units 19 to 24) 47° 50′ 32 47 Grey to pink, medium- and fine-grained equigranular granite containing many small, dark-grey and green (Units 19 and 20 occur only in the northern Fortune Bay 47a to black inclusions; 47a red felsite and fine-grained area; Unit 22 occurs only on Brunette Island) 47b granite, developed locally at pluton’s margin; 47b Red micaceous siltstone and interbedded, buff-weath- 10 pink-to brown quartz-feldspar porphyry (Red Head 24 31 Porphyry) ering, quartzitic arkose and pebble conglomerate 20′ Pink, buff-weathering, medium- to coarse-grained, Be88 OLD WOMAN STOCK 23 cross-bedded, quartzitic arkose and granule to pebble Pink, medium- and coarse-grained, porphyritic biotite 42 46 23a conglomerate; locally contains red siltstone; 23a red 32 granite; minor aplite 31 23b pebble conglomerate; 23b quartzitic arkose as in 23, MAP 98-02 GREAT BAY DE L’EAU FORMATION (Units 44 and 45) containing minor amounts of red siltstone 37 9 83 Pyr 45 Grey mafic sills and flows 22 Red and grey, thin-bedded siltstone, and fine-grained 37 GEOLOGY OF THE CONNAIGRE PENINSULA 19b sandstone and interbedded buff, coarse-grained, cross 10 25 19b Pyr 81 Red, purple and buff, pebble to boulder conglomerate; bedded quartzitic arkose; minor bright-red shale and 32 25 42 44 W,Sn 91 minor green conglomerate and red and blackshale; green-grey and black-grey and black siltstone AND ADJACENT AREAS, -

Newsletter Past President: Terry Bishop-Stirling Treasurer: Nancy Snedden
The Newfoundland Historical Society Winter 2010 1905-2010 EXECUTIVE President: Melanie Martin Vice President: Larry Coady Newsletter Past President: Terry Bishop-Stirling Treasurer: Nancy Snedden Councillors: Robert Parsons, NHS Grand Bank Regional Representative Vicki Barbour David Bradley When the Canadian Coast Guard (Alumnus), Newfoundland and Labrador division, Allan Byrne called me in November 2009 to ask if they could present me with their “Polaris” award Terry Carlson John Cheeseman on December 17, 2009, I was honored by their gesture. Not an annual award, it had Larry Dohey been given out only two previous times in the years of the Alumnus’ existence. Mekaela Mahoney The citation reads: “In Recognition of the Outstanding Contribution to the Preservation Joan Ritcey and Public Awareness of the Marine Heritage and History in the Province of Fred Smith Heidi Thorne Newfoundland and Labrador” and was presented in the Coast Guard building on St. Fran Warren John’s South Side. Jeff Webb Mike Wilkshire (Newsletter In my twenty-three years of research and writing of North Atlantic ships and sailors, Editor) survivors and victims, especially those of Newfoundland and Labrador, this is the first public recognition I’ve received for writing about the sea and shipwrecks. Regional Representatives: Meanwhile research and writing continues unabated from my keyboard here in Grand Betty Jerrett, Bay Roberts Bank. In the fall of 2009, Courting Disaster: True Crime and Mischief on Land and Sea Robert Parsons, Grand became my twenty-second book. Wrecked and Ruined: Curious Sea Tales from the Bank Eastern Edge by Pottersfield Press, Nova Scotia, is currently in press and slated for Doug Wells, Harbour Breton appearance in March/April of this year. -

Rental Housing Portfolio March 2021.Xlsx
Rental Housing Portfolio Profile by Region - AVALON - March 31, 2021 NL Affordable Housing Partner Rent Federal Community Community Housing Approved Units Managed Co-op Supplement Portfolio Total Total Housing Private Sector Non Profit Adams Cove 1 1 Arnold's Cove 29 10 39 Avondale 3 3 Bareneed 1 1 Bay Bulls 1 1 10 12 Bay Roberts 4 15 19 Bay de Verde 1 1 Bell Island 90 10 16 116 Branch 1 1 Brigus 5 5 Brownsdale 1 1 Bryants Cove 1 1 Butlerville 8 8 Carbonear 26 4 31 10 28 99 Chapel Cove 1 1 Clarke's Beach 14 24 38 Colinet 2 2 Colliers 3 3 Come by Chance 3 3 Conception Bay South 36 8 14 3 16 77 Conception Harbour 8 8 Cupids 8 8 Cupids Crossing 1 1 Dildo 1 1 Dunville 11 1 12 Ferryland 6 6 Fox Harbour 1 1 Freshwater, P. Bay 8 8 Gaskiers 2 2 Rental Housing Portfolio Profile by Region - AVALON - March 31, 2021 NL Affordable Housing Partner Rent Federal Community Community Housing Approved Units Managed Co-op Supplement Portfolio Total Total Housing Goobies 2 2 Goulds 8 4 12 Green's Harbour 2 2 Hant's Harbour 0 Harbour Grace 14 2 6 22 Harbour Main 1 1 Heart's Content 2 2 Heart's Delight 3 12 15 Heart's Desire 2 2 Holyrood 13 38 51 Islingston 2 2 Jerseyside 4 4 Kelligrews 24 24 Kilbride 1 24 25 Lower Island Cove 1 1 Makinsons 2 1 3 Marysvale 4 4 Mount Carmel-Mitchell's Brook 2 2 Mount Pearl 208 52 18 10 24 28 220 560 New Harbour 1 10 11 New Perlican 0 Norman's Cove-Long Cove 5 12 17 North River 4 1 5 O'Donnels 2 2 Ochre Pit Cove 1 1 Old Perlican 1 8 9 Paradise 4 14 4 22 Placentia 28 2 6 40 76 Point Lance 0 Port de Grave 0 Rental Housing Portfolio Profile by Region - AVALON - March 31, 2021 NL Affordable Housing Partner Rent Federal Community Community Housing Approved Units Managed Co-op Supplement Portfolio Total Total Housing Portugal Cove/ St. -
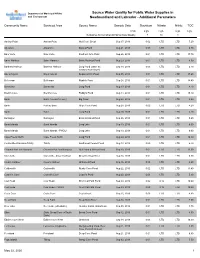
Source Water Quality for Public Water Supplies in Newfoundland And
Department of Municipal Affairs Source Water Quality for Public Water Supplies in and Environment Newfoundland and Labrador - Additional Parameters Community Name Serviced Area Source Name Sample Date Strontium Nitrate Nitrite TOC Units mg/L mg/L mg/L mg/L Guidelines for Canadian Drinking Water Quality 7 10 1 Anchor Point Anchor Point Well Cove Brook Sep 17, 2019 0.02 LTD LTD 7.20 Aquaforte Aquaforte Davies Pond Aug 21, 2019 0.00 LTD LTD 6.30 Baie Verte Baie Verte Southern Arm Pond Sep 26, 2019 0.01 LTD LTD 17.70 Baine Harbour Baine Harbour Baine Harbour Pond Aug 29, 2019 0.01 LTD LTD 9.50 Bartletts Harbour Bartletts Harbour Long Pond (same as Sep 18, 2019 0.03 LTD LTD 6.70 Castors River North) Bay L'Argent Bay L'Argent Sugarloaf Hill Pond Sep 05, 2019 0.01 LTD LTD 17.60 Belleoram Belleoram Rabbits Pond Sep 24, 2019 0.01 LTD LTD 14.40 Bonavista Bonavista Long Pond Aug 13, 2019 0.01 LTD LTD 4.10 Brent's Cove Brent's Cove Paddy's Pond Aug 14, 2019 0.01 LTD LTD 15.10 Burin Burin (+Lewin's Cove) Big Pond Aug 28, 2019 0.01 LTD LTD 4.90 Burin Port au Bras Gripe Cove Pond Aug 28, 2019 0.02 LTD LTD 4.20 Burin Burin Long Pond Aug 28, 2019 0.01 LTD LTD 4.10 Burlington Burlington Eastern Island Pond Sep 26, 2019 0.01 LTD LTD 9.60 Burnt Islands Burnt Islands Long Lake Sep 10, 2019 0.01 LTD LTD 6.00 Burnt Islands Burnt Islands - PWDU Long Lake Sep 10, 2019 0.01 LTD LTD 6.00 Cape Freels North Cape Freels North Long Pond Aug 20, 2019 0.01 LTD LTD 10.30 Centreville-Wareham-Trinity Trinity Southwest Feeder Pond Aug 13, 2019 0.00 LTD LTD 6.70 Channel-Port -

19 Century Newfoundland Outport Merchants the Jersey Room, Burin
19th century Newfoundland outport merchants The Jersey Room, Burin, c. 1885, S.H. Parsons photo (GPA collection). submitted to Provincial Historic Commemorations Program Dept. Business, Tourism, Culture & Rural Development P.O. Box 8700 St. John's, NL A1E 1J3 submitted by Robert H. Cuff Historian/Writer Gerald Penney Associates Limited PO Box 428, St. John’s, NL A1C 5K4 10 November 2014 Executive Summary In their impact on Newfoundland and Labrador’s economic development, patterns of settlement, and community life, 19th century outport merchants made a significant historic contribution. Their secondary impact, on the Province’s political and cultural development, may be less obvious but was nonetheless vital. Each merchant had a demonstrable impact beyond his home community, in that each supplied nearby communities. Although a merchant’s commercial home sphere was typically in the headquarters bay or region, the majority of the outport merchants were also involved in both fishing and in supplying planters/ fishers in migratory or vessel-based fisheries elsewhere: the Labrador and French Shore fisheries; the seal hunt; and the western boat and Bank fisheries of the south coast. For the purposes of this review it was found helpful to draw a distinction between “resident outport merchants” who lived the full range of their adult lives in rural Newfoundland and the “merchant gentry” whose outport residency was an episode in their business and family life which was otherwise substantially spent in the Old Country or in St. John’s. The resident group may be more worthy of consideration for the Province’s commemoration program. Existing commemorations tend to favour the merchant gentry. -

ERIC COBHAM: the Pirate Who Never Was?
- ERIC COBHAM: The Pirate Who Never Was? BY OLAF U JANZEN GRENFELL CAMPUS, MEMORIAL UNIVERSITY THE HISTORY OF NEWFOUNDLAND AND his article focuses on another “certainty” among LABRADOR IS FULL OF LEGENDS AND some, namely the life and career of the pirate Eric Cobham who, with his wife Maria Lindsay, MYTHS THAT ARE BASED MORE ON BELIEF Tallegedly established a base at Sandy Point in Bay THAN ON EVIDENCE. THUS MANY PEOPLE St George. The two, for 20 years or so, allegedly “KNOW” THAT IN 1497, CABOT MADE attacked and plundered ships and vessels as they made their way across the Gulf of St Lawrence, and murdered LANDFALL AT BONAVISTA, AND THAT their crews. I will not attempt to explain why people are THE BEOTHUK PEOPLE WERE HUNTED so determined to believe that Cobham existed–I’ll leave FOR SPORT. SOME PEOPLE BELIEVE THAT that to others. Instead I’ll focus on the reasons why the Cobham story is so improbable. This is not to say that SHEILA NAGEIRA WAS A YOUNG IRISH pirates didn’t operate in Early Modern Newfoundland. NOBLEWOMAN RESCUED BY PETER They did. This article will begin there, with an EASTON FROM DUTCH PRIVATEERS IN examination of the reality of piracy in Newfoundland, before I turn my attention to the lack of reality of THE EARLY 1600s, WHEREUPON SHE FELL Eric Cobham. IN LOVE WITH GILBERT PIKE, ONE OF EASTON’S LIEUTENANTS, SUBSEQUENTLY Piracy in the 16th and 17th centuries MARRIED, AND EVENTUALLY SETTLED In 1604, the owners of the Hopewell of London prepared their vessel for a voyage to the Newfoundland fishery IN CARBONEAR.