Chromatin-Associated HMGA and HMGB Proteins: Versatile Co-Regulators of DNA-Dependent Processes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Functional Compensation Among HMGN Variants Modulates the Dnase I Hypersensitive Sites at Enhancers
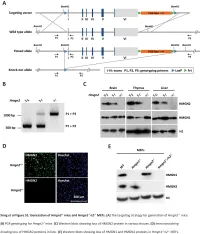
Downloaded from genome.cshlp.org on October 9, 2021 - Published by Cold Spring Harbor Laboratory Press Research Functional compensation among HMGN variants modulates the DNase I hypersensitive sites at enhancers Tao Deng,1,12 Z. Iris Zhu,2,12 Shaofei Zhang,1 Yuri Postnikov,1 Di Huang,2 Marion Horsch,3 Takashi Furusawa,1 Johannes Beckers,3,4,5 Jan Rozman,3,5 Martin Klingenspor,6,7 Oana Amarie,3,8 Jochen Graw,3,8 Birgit Rathkolb,3,5,9 Eckhard Wolf,9 Thure Adler,3 Dirk H. Busch,10 Valérie Gailus-Durner,3 Helmut Fuchs,3 Martin Hrabeˇ de Angelis,3,4,5 Arjan van der Velde,2,13 Lino Tessarollo,11 Ivan Ovcherenko,2 David Landsman,2 and Michael Bustin1 1Protein Section, Laboratory of Metabolism, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland 20892, USA; 2Computational Biology Branch, National Center for Biotechnology Information, National Library of Medicine, Bethesda, Maryland 20892, USA; 3German Mouse Clinic, Institute of Experimental Genetics, Helmholtz Zentrum München, German Research Center for Environmental Health, 85764 Neuherberg, Germany; 4Experimental Genetics, Center of Life and Food Sciences Weihenstephan, Technische Universität München, 85354 Freising-Weihenstephan, Germany; 5German Center for Diabetes Research (DZD), 85764 Neuherberg, Germany; 6Molecular Nutritional Medicine, Technische Universität München, 85350 Freising, Germany; 7Center for Nutrition and Food Sciences, Technische Universität München, 85350 Freising, Germany; 8Institute of Developmental Genetics (IDG), 85764 -

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -

Of Keeping and Tipping the Balance: Host Regulation and Viral Modulation of IRF3-Dependent IFNB1 Expression
viruses Review Of Keeping and Tipping the Balance: Host Regulation and Viral Modulation of IRF3-Dependent IFNB1 Expression Hella Schwanke 1,2 , Markus Stempel 1,2 and Melanie M. Brinkmann 1,2,* 1 Institute of Genetics, Technische Universität Braunschweig, 38106 Braunschweig, Germany; [email protected] (H.S.); [email protected] (M.S.) 2 Viral Immune Modulation Research Group, Helmholtz Centre for Infection Research, 38124 Braunschweig, Germany * Correspondence: [email protected]; Tel.: +49-531-6181-3069 Received: 15 June 2020; Accepted: 3 July 2020; Published: 7 July 2020 Abstract: The type I interferon (IFN) response is a principal component of our immune system that allows to counter a viral attack immediately upon viral entry into host cells. Upon engagement of aberrantly localised nucleic acids, germline-encoded pattern recognition receptors convey their find via a signalling cascade to prompt kinase-mediated activation of a specific set of five transcription factors. Within the nucleus, the coordinated interaction of these dimeric transcription factors with coactivators and the basal RNA transcription machinery is required to access the gene encoding the type I IFN IFNβ (IFNB1). Virus-induced release of IFNβ then induces the antiviral state of the system and mediates further mechanisms for defence. Due to its key role during the induction of the initial IFN response, the activity of the transcription factor interferon regulatory factor 3 (IRF3) is tightly regulated by the host and fiercely targeted by viral proteins at all conceivable levels. In this review, we will revisit the steps enabling the trans-activating potential of IRF3 after its activation and the subsequent assembly of the multi-protein complex at the IFNβ enhancer that controls gene expression. -

HMGB1 in Health and Disease R
Donald and Barbara Zucker School of Medicine Journal Articles Academic Works 2014 HMGB1 in health and disease R. Kang R. C. Chen Q. H. Zhang W. Hou S. Wu See next page for additional authors Follow this and additional works at: https://academicworks.medicine.hofstra.edu/articles Part of the Emergency Medicine Commons Recommended Citation Kang R, Chen R, Zhang Q, Hou W, Wu S, Fan X, Yan Z, Sun X, Wang H, Tang D, . HMGB1 in health and disease. 2014 Jan 01; 40():Article 533 [ p.]. Available from: https://academicworks.medicine.hofstra.edu/articles/533. Free full text article. This Article is brought to you for free and open access by Donald and Barbara Zucker School of Medicine Academic Works. It has been accepted for inclusion in Journal Articles by an authorized administrator of Donald and Barbara Zucker School of Medicine Academic Works. Authors R. Kang, R. C. Chen, Q. H. Zhang, W. Hou, S. Wu, X. G. Fan, Z. W. Yan, X. F. Sun, H. C. Wang, D. L. Tang, and +8 additional authors This article is available at Donald and Barbara Zucker School of Medicine Academic Works: https://academicworks.medicine.hofstra.edu/articles/533 NIH Public Access Author Manuscript Mol Aspects Med. Author manuscript; available in PMC 2015 December 01. NIH-PA Author ManuscriptPublished NIH-PA Author Manuscript in final edited NIH-PA Author Manuscript form as: Mol Aspects Med. 2014 December ; 0: 1–116. doi:10.1016/j.mam.2014.05.001. HMGB1 in Health and Disease Rui Kang1,*, Ruochan Chen1, Qiuhong Zhang1, Wen Hou1, Sha Wu1, Lizhi Cao2, Jin Huang3, Yan Yu2, Xue-gong Fan4, Zhengwen Yan1,5, Xiaofang Sun6, Haichao Wang7, Qingde Wang1, Allan Tsung1, Timothy R. -
Supplemental Data.Pdf
Table S1. Summary of sequencing results A. DNase‐seq data % Align % Mismatch % >=Q30 bases Sample ID # Reads % unique reads (PF) Rate (PF) (PF) WT_1 84,301,522 86.76 0.52 93.34 96.30 WT_2 98,744,222 84.97 0.51 93.75 89.94 Hmgn1‐/‐_1 79,620,656 83.94 0.86 90.58 88.65 Hmgn1‐/‐_2 62,673,782 84.13 0.87 91.30 89.18 Hmgn2‐/‐_1 87,734,440 83.49 0.71 91.81 90.00 Hmgn2‐/‐_2 82,498,808 83.25 0.69 92.73 90.66 Hmgn1‐/‐n2‐/‐_1 71,739,638 68.51 2.31 81.11 89.22 Hmgn1‐/‐n2‐/‐_2 74,113,682 68.19 2.37 81.16 86.57 B. ChIP‐seq data Histone % Align % Mismatch % >=Q30 % unique Genotypes # Reads marks (PF) Rate (PF) bases (PF) reads H3K4me1 100670054 92.99 0.28 91.21 87.29 H3K4me3 67064272 91.97 0.35 89.11 27.15 WT H3K27ac 90,340,242 93.57 0.28 95.02 89.80 input 111,292,572 78.24 0.55 96.07 86.99 H3K4me1 84598176 92.34 0.33 91.2 81.69 H3K4me3 90032064 92.19 0.44 88.76 15.81 Hmgn1‐/‐n2‐/‐ H3K27ac 86,260,526 93.40 0.29 94.94 87.49 input 78,142,334 78.47 0.56 95.82 81.11 C. MNase‐seq data % Mismatch % >=Q30 bases % unique Sample ID # Reads % Align (PF) Rate (PF) (PF) reads WT_1_Extensive 45,232,694 55.23 1.49 90.22 81.73 WT_1_Limited 105,460,950 58.03 1.39 90.81 79.62 WT_2_Extensive 40,785,338 67.34 1.06 89.76 89.60 WT_2_Limited 105,738,078 68.34 1.05 90.29 85.96 Hmgn1‐/‐n2‐/‐_1_Extensive 117,927,050 55.74 1.49 89.50 78.01 Hmgn1‐/‐n2‐/‐_1_Limited 61,846,742 63.76 1.22 90.57 84.55 Hmgn1‐/‐n2‐/‐_2_Extensive 137,673,830 60.04 1.30 89.28 78.99 Hmgn1‐/‐n2‐/‐_2_Limited 45,696,614 62.70 1.21 90.71 85.52 D. -

Genomic Interaction Between ER and HMGB2 Identifies DDX18 As A
Oncogene (2015) 34, 3871–3880 © 2015 Macmillan Publishers Limited All rights reserved 0950-9232/15 www.nature.com/onc ORIGINAL ARTICLE Genomic interaction between ER and HMGB2 identifies DDX18 as a novel driver of endocrine resistance in breast cancer cells AM Redmond1,2, C Byrne1, FT Bane1, GD Brown2, P Tibbitts1,KO’Brien1, ADK Hill1, JS Carroll2 and LS Young1 Breast cancer resistance to endocrine therapies such as tamoxifen and aromatase inhibitors is a significant clinical problem. Steroid receptor coactivator-1 (SRC-1), a coregulatory protein of the oestrogen receptor (ER), has previously been shown to have a significant role in the progression of breast cancer. The chromatin protein high mobility group box 2 (HMGB2) was identified as an SRC-1 interacting protein in the endocrine-resistant setting. We investigated the expression of HMGB2 in a cohort of 1068 breast cancer patients and found an association with increased disease-free survival time in patients treated with endocrine therapy. However, it was also verified that HMGB2 expression could be switched on in endocrine-resistant tumours from breast cancer patients. To explore the function of this poorly characterized protein, we performed HMGB2 ChIPseq and found distinct binding patterns between the two contexts. In the resistant setting, the HMGB2, SRC-1 and ER complex are enriched at promoter regions of target genes, with bioinformatic analysis indicating a switch in binding partners between the sensitive and resistant phenotypes. Integration of binding and gene expression data reveals a concise set of target genes of this complex including the RNA helicase DDX18. Modulation of DDX18 directly affects growth of tamoxifen-resistant cells, suggesting that it may be a critical downstream effector of the HMGB2:ER complex. -

A Dissertation Entitled Alpha7 Nicotinic
A Dissertation entitled Alpha7 Nicotinic Acetylcholine Receptor: Novel Role in Macrophage Survival and Murine Atherosclerosis by Robert H. Lee Submitted to the Graduate Faculty as partial fulfillment of the requirements for the Doctor of Philosophy Degree in Biomedical Sciences _________________________________________ Guillermo Vazquez, PhD, Committee Chair _________________________________________ David Giovannucci, PhD, Committee Member _________________________________________ Joseph Margiotta, PhD, Committee Member _________________________________________ Sandrine Pierre, PhD, Committee Member _________________________________________ R. Mark Wooten, PhD, Committee Member _________________________________________ Patricia R. Komuniecki, PhD, Dean College of Graduate Studies The University of Toledo December 2014 Copyright 2014, Robert Hugh Lee This document is copyrighted material. Under copyright law, no parts of this document may be reproduced without the expressed permission of the author. An Abstract of Alpha7 Nicotinic Acetylcholine Receptor: Novel Role in Macrophage Survival and Murine Atherosclerosis by Robert H. Lee Submitted to the Graduate Faculty as partial fulfillment of the requirements for the Doctor of Philosophy Degree in Biomedical Sciences The University of Toledo December 2014 Atherosclerosis is a chronic inflammatory disease, characterized by infiltration and accumulation of leukocytes within the vascular wall. Macrophages play a particularly crucial role in all stages of atherosclerotic lesion development. These -

GATA2 Regulates Mast Cell Identity and Responsiveness to Antigenic Stimulation by Promoting Chromatin Remodeling at Super- Enhancers
ARTICLE https://doi.org/10.1038/s41467-020-20766-0 OPEN GATA2 regulates mast cell identity and responsiveness to antigenic stimulation by promoting chromatin remodeling at super- enhancers Yapeng Li1, Junfeng Gao 1, Mohammad Kamran1, Laura Harmacek2, Thomas Danhorn 2, Sonia M. Leach1,2, ✉ Brian P. O’Connor2, James R. Hagman 1,3 & Hua Huang 1,3 1234567890():,; Mast cells are critical effectors of allergic inflammation and protection against parasitic infections. We previously demonstrated that transcription factors GATA2 and MITF are the mast cell lineage-determining factors. However, it is unclear whether these lineage- determining factors regulate chromatin accessibility at mast cell enhancer regions. In this study, we demonstrate that GATA2 promotes chromatin accessibility at the super-enhancers of mast cell identity genes and primes both typical and super-enhancers at genes that respond to antigenic stimulation. We find that the number and densities of GATA2- but not MITF-bound sites at the super-enhancers are several folds higher than that at the typical enhancers. Our studies reveal that GATA2 promotes robust gene transcription to maintain mast cell identity and respond to antigenic stimulation by binding to super-enhancer regions with dense GATA2 binding sites available at key mast cell genes. 1 Department of Immunology and Genomic Medicine, National Jewish Health, Denver, CO 80206, USA. 2 Center for Genes, Environment and Health, National Jewish Health, Denver, CO 80206, USA. 3 Department of Immunology and Microbiology, University of Colorado Anschutz Medical Campus, Aurora, ✉ CO 80045, USA. email: [email protected] NATURE COMMUNICATIONS | (2021) 12:494 | https://doi.org/10.1038/s41467-020-20766-0 | www.nature.com/naturecommunications 1 ARTICLE NATURE COMMUNICATIONS | https://doi.org/10.1038/s41467-020-20766-0 ast cells (MCs) are critical effectors in immunity that at key MC genes. -

P53 Promotes Inflammation-Associated
Research Article p53 promotes inflammation-associated hepatocarcinogenesis by inducing HMGB1 release ⇑ He-Xin Yan1,2, , Hong-Ping Wu1,2, Hui-Lu Zhang1, Charles Ashton2, Chang Tong2, Han Wu1, ⇑ Qi-Jun Qian1, Hong-Yang Wang1, Qi-Long Ying2, 1Eastern Hepatobiliary Surgery Hospital, Second Military Medical University, Shanghai 200433, China; 2Eli and Edythe Broad Center for Regenerative Medicine and Stem Cell Research at USC, Department of Cell and Neurobiology, Keck School of Medicine, University of Southern California, Los Angeles, CA 90033, USA Background & Aims: Hepatocellular carcinoma (HCC) develops Ó 2013 European Association for the Study of the Liver. Published in response to chronic hepatic injury. Although induced cell death by Elsevier B.V. All rights reserved. is regarded as the major component of p53 tumor-suppressive activity, we recently found that sustained p53 activation subse- quent to DNA damage promotes inflammation-associated hepatocarcinogenesis. Here we aim at exploring the mechanism Introduction linking p53 activation and hepatic inflammation during hepatocarcinogenesis. A strong association between chronic tissue injury and tumori- Methods: p53À/À hepatocytes expressing inducible p53 and pri- genesis has been established by many clinical data [1–3]. The mary wild type hepatocytes were treated to induce p53 expres- mammalian liver is vulnerable to damage induced by toxic chem- sion. The supernatants were collected and analyzed for the icals and hepatitis viruses, all of which increase the risk of hepa- presence of released inflammatory cytokines. Ethyl pyruvate tocellular carcinoma (HCC). Chronic hepatic inflammation, as a was used in a rat model of carcinogen-induced hepatocarcino- consequence of chronic injury, frequently precedes the develop- genesis to examine its effect on p53-dependent chronic hepatic ment of HCC [4]. -

Deficiency of the Novel High Mobility Group Protein HMGXB4 Protects Against Systemic Inflammation-Induced Endotoxemia in Mice
Deficiency of the novel high mobility group protein HMGXB4 protects against systemic inflammation-induced endotoxemia in mice Xiangqin Hea,b,c,1, Kunzhe Dongc,1, Jian Shenc,d, Guoqing Huc, Jinhua Liuc,e, Xiuhua Kangc,e, Liang Wangf, Reem T. Atawiac, Islam Osmanc, Robert W. Caldwellc, Meixiang Xiangd, Wei Zhange, Zeqi Zhengf, Liwu Lig, David J. R. Fultonc,h, Keyu Denga,b, Hongbo Xina,b,2, and Jiliang Zhouc,2 aThe National Engineering Research Center for Bioengineering Drugs and Technologies, The Institute of Translational Medicine, Nanchang University, 330031 Nanchang, Jiangxi, China; bSchool of Life Sciences, Nanchang University, 330031 Nanchang, Jiangxi, China; cDepartment of Pharmacology and Toxicology, Medical College of Georgia, Augusta University, Augusta, GA 30912; dDepartment of Cardiology, The Second Affiliated Hospital, Zhejiang University School of Medicine, 310009 Hangzhou, China; eDepartment of Respiratory Medicine, The First Affiliated Hospital of Nanchang University, 330006 Nanchang, Jiangxi, China; fDepartment of Cardiology, The First Affiliated Hospital of Nanchang University, 330006 Nanchang, Jiangxi, China; gDepartment of Biological Sciences, Virginia Polytechnic Institute and State University, Blacksburg, VA 24061-0910; and hVascular Biology Center, Medical College of Georgia, Augusta University, Augusta, GA 30912 Edited by Kevin J. Tracey, Feinstein Institute for Medical Research, Manhasset, NY, and accepted by Editorial Board Member Carl F. Nathan December 16, 2020 (received for review October 26, 2020) Sepsis is a major cause of mortality in intensive care units, which Escherichia coli (4). LPS induces a systemic inflammatory re- results from a severely dysregulated inflammatory response that sponse via the pattern recognition receptor CD14 and Toll-like ultimately leads to organ failure. -

Itraq‑Based Proteomic Analysis of Endotoxin Tolerance Induced by Lipopolysaccharide
584 MOLECULAR MEDICINE REPORTS 20: 584-592, 2019 iTRAQ‑based proteomic analysis of endotoxin tolerance induced by lipopolysaccharide QIAN ZHANG1, YINGCHUN HU2, JING ZHANG1 and CUNLIANG DENG1 Departments of 1Infectious Diseases and 2Emergency, The Affiliated Hospital of Southwest Medical University, Luzhou, Sichuan 646000, P.R. China Received August 25, 2018; Accepted February 15, 2019 DOI: 10.3892/mmr.2019.10264 Abstract. The purpose of the present study was to investigate Introduction the differentially expressed proteins between endotoxin toler- ance and sepsis. Cell models of an endotoxin tolerance group Sepsis is a serious healthcare concern due to its high costs (ET group) and sepsis group [lipopolysaccharide (LPS) group] and mortality rate (1). It is triggered mainly by Gram-negative were established using LPS and evaluated using ELISA and bacteria (2) and lipopolysaccharides (LPS) are the main flow cytometry methods. Differentially expressed proteins component of the outer membrane of Gram-negative bacteria. between the ET and the LPS groups were identified using Previous studies have demonstrated that LPS serve an important isobaric tags for relative and absolute quantitation (iTRAQ) role in sepsis, and can stimulate the mononuclear phagocytic analysis and evaluated by bioinformatics analysis. The expres- system to produce a large number of inflammatory factors sion of core proteins was detected by western blotting. It was and cause systemic inflammatory response, sepsis, infectious identified that the expression of tumor necrosis factor‑α and shock and multiple organ dysfunctions (3). It has been reported interleukin-6 was significantly decreased in the ET group that following low-dose LPS stimulation, the host can survive a compared with the LPS group. -

HMGB2 Polyclonal Antibody Catalog Number:14597-1-AP Featured Product 8 Publications
For Research Use Only HMGB2 Polyclonal antibody Catalog Number:14597-1-AP Featured Product 8 Publications www.ptglab.com Catalog Number: GenBank Accession Number: Purification Method: Basic Information 14597-1-AP BC001063 Antigen affinity purification Size: GeneID (NCBI): Recommended Dilutions: 150ul , Concentration: 900 μg/ml by 3148 WB 1:500-1:3000 Nanodrop and 400 μg/ml by Bradford Full Name: IP 0.5-4.0 ug for IP and 1:500-1:1000 method using BSA as the standard; high-mobility group box 2 for WB Source: IHC 1:50-1:500 Calculated MW: IF 1:50-1:500 Rabbit 24 kDa Isotype: Observed MW: IgG 33-35 kDa Immunogen Catalog Number: AG6135 Applications Tested Applications: Positive Controls: IF, IHC, IP, WB,ELISA WB : Jurkat cells, HEK-293 cells, HL-60 cells, human Cited Applications: cerebellum tissue, K-562 cells WB IP : HEK-293 cells, Species Specificity: IHC : mouse brain tissue, human, mouse, rat Cited Species: IF : HepG2 cells, human, mouse, rat Note-IHC: suggested antigen retrieval with TE buffer pH 9.0; (*) Alternatively, antigen retrieval may be performed with citrate buffer pH 6.0 High mobility group protein B2 (HMGB2) belongs to a family of highly conserved proteins that contain HMG box Background Information domains (11246022,14871457). All three family members (HMGB1, HMGB2, and HMGB3) contain two HMG box domains and a C-terminal acidic domain. HMGB1 is a widely expressed and highly abundant protein (14871457). HMGB2 is widely expressed during embryonic development, but it is restricted to lymphoid organs and testis in adult animals (11262228). HMGB3 is only expressed during embryogenesis (9598312).