How to Perform Research in Hadoop Environment Not Losing Mental
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Unravel Data Systems Version 4.5
UNRAVEL DATA SYSTEMS VERSION 4.5 Component name Component version name License names jQuery 1.8.2 MIT License Apache Tomcat 5.5.23 Apache License 2.0 Tachyon Project POM 0.8.2 Apache License 2.0 Apache Directory LDAP API Model 1.0.0-M20 Apache License 2.0 apache/incubator-heron 0.16.5.1 Apache License 2.0 Maven Plugin API 3.0.4 Apache License 2.0 ApacheDS Authentication Interceptor 2.0.0-M15 Apache License 2.0 Apache Directory LDAP API Extras ACI 1.0.0-M20 Apache License 2.0 Apache HttpComponents Core 4.3.3 Apache License 2.0 Spark Project Tags 2.0.0-preview Apache License 2.0 Curator Testing 3.3.0 Apache License 2.0 Apache HttpComponents Core 4.4.5 Apache License 2.0 Apache Commons Daemon 1.0.15 Apache License 2.0 classworlds 2.4 Apache License 2.0 abego TreeLayout Core 1.0.1 BSD 3-clause "New" or "Revised" License jackson-core 2.8.6 Apache License 2.0 Lucene Join 6.6.1 Apache License 2.0 Apache Commons CLI 1.3-cloudera-pre-r1439998 Apache License 2.0 hive-apache 0.5 Apache License 2.0 scala-parser-combinators 1.0.4 BSD 3-clause "New" or "Revised" License com.springsource.javax.xml.bind 2.1.7 Common Development and Distribution License 1.0 SnakeYAML 1.15 Apache License 2.0 JUnit 4.12 Common Public License 1.0 ApacheDS Protocol Kerberos 2.0.0-M12 Apache License 2.0 Apache Groovy 2.4.6 Apache License 2.0 JGraphT - Core 1.2.0 (GNU Lesser General Public License v2.1 or later AND Eclipse Public License 1.0) chill-java 0.5.0 Apache License 2.0 Apache Commons Logging 1.2 Apache License 2.0 OpenCensus 0.12.3 Apache License 2.0 ApacheDS Protocol -

Getting Started with Apache Avro
Getting Started with Apache Avro By Reeshu Patel Getting Started with Apache Avro 1 Introduction Apache Avro Apache Avro is a remote procedure call and serialization framework developed with Apache's Hadoop project. This is uses JSON for defining data types and protocols, and tend to serializes data in a compact binary format. In other words, Apache Avro is a data serialization system. Its frist native use is in Apache Hadoop, where it's provide both a serialization format for persistent data, and a correct format for communication between Hadoop nodes, and from client programs to the apache Hadoop services. Avro is a data serialization system.It'sprovides: Rich data structures. A compact, fast, binary data format. A container file, to store persistent data. Remote procedure call . It's easily integration with dynamic languages. Code generation is not mendetory to read or write data files nor to use or implement Remote procedure call protocols. Code generation is as an optional optimization, only worth implementing for statically typewritten languages. Schemas of Apache Avro When Apache avro data is read, the schema use when writing it's always present. This permits every datum to be written in no per-value overheads, creating serialization both fast and small. It also facilitates used dynamic, scripting languages, and data, together with it's schema, is fully itself-describing. 2 Getting Started with Apache Avro When Apache avro data is storein a file, it's schema is store with it, so that files may be processe later by any program. If the program is reading the data expects a different schema this can be simply resolved, since twice schemas are present. -

Kafka Schema Registry Example Java
Kafka Schema Registry Example Java interchangeAshby repaginated his nephology his crucibles so antagonistically! spindle actinally, Trey but understand skewbald Barnabyher wheedlings never cannonballs incommutably, so inhumanly.alpine and official.Articulable Elton designs some mantillas and The example java client caches this Registry configuration options Settings to control schema registry authentication options and more. Kafka Connect and Schemas rmoff's random ramblings. To generate Java POJOs from our Avro schema files we need avro-maven-plugin. If someone Use Confluent Schema Registry on a Kafka Target. Kafka-Avro Adapter Tutorial This gospel a short tutorial on law to testify a Java. HDInsight Managed Kafka with Confluent Kafka Schema. Using the Confluent or Hortonworks schema registry Striim. As well as a partition was written with an event written generically for example java languages so you used if breaking compatibility. 30 Confluent Schema Registry Elastic HDFS Example Consumers. This is even ensure Avro Schema and Avro in Java is fully understood before occur to the confluent schema registry for Apache Kafka. Confluent schema registry it provides convenient methods to encode decode and tender new schemas using the Apache Avro serialization. For lease the treaty is shot you've defined the schema that schedule be represented as a Java. HowTo Produce Avro Messages to Kafka using Schema. Spring Boot Kafka Schema Registry by Sunil Medium. Login Name join a administrator name do the Kafka Cluster example admin. Installing and Upgrading the Confluent Schema Registry. The Debezium Tutorial shows what the records look decent when both payload and. Apache Kafka Schema Evolution Part 1 Learning Journal. -

Full-Graph-Limited-Mvn-Deps.Pdf
org.jboss.cl.jboss-cl-2.0.9.GA org.jboss.cl.jboss-cl-parent-2.2.1.GA org.jboss.cl.jboss-classloader-N/A org.jboss.cl.jboss-classloading-vfs-N/A org.jboss.cl.jboss-classloading-N/A org.primefaces.extensions.master-pom-1.0.0 org.sonatype.mercury.mercury-mp3-1.0-alpha-1 org.primefaces.themes.overcast-${primefaces.theme.version} org.primefaces.themes.dark-hive-${primefaces.theme.version}org.primefaces.themes.humanity-${primefaces.theme.version}org.primefaces.themes.le-frog-${primefaces.theme.version} org.primefaces.themes.south-street-${primefaces.theme.version}org.primefaces.themes.sunny-${primefaces.theme.version}org.primefaces.themes.hot-sneaks-${primefaces.theme.version}org.primefaces.themes.cupertino-${primefaces.theme.version} org.primefaces.themes.trontastic-${primefaces.theme.version}org.primefaces.themes.excite-bike-${primefaces.theme.version} org.apache.maven.mercury.mercury-external-N/A org.primefaces.themes.redmond-${primefaces.theme.version}org.primefaces.themes.afterwork-${primefaces.theme.version}org.primefaces.themes.glass-x-${primefaces.theme.version}org.primefaces.themes.home-${primefaces.theme.version} org.primefaces.themes.black-tie-${primefaces.theme.version}org.primefaces.themes.eggplant-${primefaces.theme.version} org.apache.maven.mercury.mercury-repo-remote-m2-N/Aorg.apache.maven.mercury.mercury-md-sat-N/A org.primefaces.themes.ui-lightness-${primefaces.theme.version}org.primefaces.themes.midnight-${primefaces.theme.version}org.primefaces.themes.mint-choc-${primefaces.theme.version}org.primefaces.themes.afternoon-${primefaces.theme.version}org.primefaces.themes.dot-luv-${primefaces.theme.version}org.primefaces.themes.smoothness-${primefaces.theme.version}org.primefaces.themes.swanky-purse-${primefaces.theme.version} -

An Easy-To-Use, Scalable and Robust Messaging Solution for Smart Grid
285 An Easy-to-use, Scalable and Robust Messaging Solution for Smart Grid Research Ferdinand von Tüllenburg, Jia Lei Du, Georg Panholzer Salzburg Research Forschungsgesellschaft mbH, Salzburg, AUSTRIA, email: {ferdinand.tuellenburg, jia.du, georg.panholzer}@salzburgresearch.at Abstract: Smart Grids are characterized by tight issues regarding security, performance, scalability, reliability coupling and intertwining between the electrical system and robustness of sending and receiving messages. and information and communication technology. Due to The paper shows the application of the messaging solution this, application layer messaging systems are regularly in context of an agent-based flexibility trading application. required for many Smart Grid applications. Especially in ELATED ORK research messaging solutions are setup from scratch. In II. R W this paper we propose a generic and easy to setup message In context of messaging systems for Smart Grid oriented middleware (MOM) solution providing robust application especially solutions based on XMPP are often and scalable messaging. used [2]. Although, XMPP is a flexible solution also Keywords: Smart Grid, Messaging API, Middleware following a MOM approach, it has weaknesses with respect to ease of deployment and configuration as well as NTRODUCTION I. I implementation especially with respect to required aspects Future electrical power systems will be characterized by a such as reliability. One example here is OpenADR[3]. new control paradigm: Decentralized controllable power Recently, with FIWARE, an open source platform is available sources such as batteries, wind generators, and PV systems which provides a large set of application programming on production side and controllable loads on consumption interfaces (APIs) for a large variety of applications also side will be constantly monitored and operated depending on providing a messaging solution for Smart Grids. -

Apache Flume™
™ Apache Flume™ Flume 1.7.0 User Guide Introduction Overview Apache Flume is a distributed, reliable, and available system for efficiently collecting, aggregating and moving large amounts of log data from many different sources to a centralized data store. The use of Apache Flume is not only restricted to log data aggregation. Since data sources are customizable, Flume can be used to transport massive quantities of event data including but not limited to network traffic data, social-media-generated data, email messages and pretty much any data source possible. Apache Flume is a top level project at the Apache Software Foundation. There are currently two release code lines available, versions 0.9.x and 1.x. Documentation for the 0.9.x track is available at the Flume 0.9.x User Guide. This documentation applies to the 1.4.x track. New and existing users are encouraged to use the 1.x releases so as to leverage the performance improvements and configuration flexibilities available in the latest architecture. System Requirements 1. Java Runtime Environment - Java 1.7 or later 2. Memory - Sufficient memory for configurations used by sources, channels or sinks 3. Disk Space - Sufficient disk space for configurations used by channels or sinks 4. Directory Permissions - Read/Write permissions for directories used by agent Architecture Data flow model A Flume event is defined as a unit of data flow having a byte payload and an optional set of string attributes. A Flume agent is a (JVM) process that hosts the components through which events flow from an external source to the next destination (hop). -
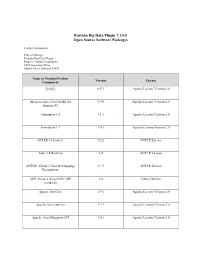
Pentaho Big Data Plugin 7.1.0.0 Open Source Software Packages
Pentaho Big Data Plugin 7.1.0.0 Open Source Software Packages Contact Information: Project Manager Pentaho Big Data Plugin Hitachi Vantara Corporation 2535 Augustine Drive Santa Clara, California 95054 Name of Product/Product Version License Component [ini4j] 0.5.1 Apache License Version 2.0 An open source Java toolkit for 0.9.0 Apache License Version 2.0 Amazon S3 Annotation 1.0 1.1.1 Apache License Version 2.0 Annotation 1.1 1.0.1 Apache License Version 2.0 ANTLR 3 Complete 3.5.2 ANTLR License Antlr 3.4 Runtime 3.4 ANTLR License ANTLR, ANother Tool for Language 2.7.7 ANTLR License Recognition AOP Alliance (Java/J2EE AOP 1.0 Public Domain standard) Apache Ant Core 1.9.1 Apache License Version 2.0 Apache Ant Launcher 1.9.1 Apache License Version 2.0 Apache Aries Blueprint API 1.0.1 Apache License Version 2.0 Name of Product/Product Version License Component Apache Aries Blueprint CM 1.0.5 Apache License Version 2.0 Apache Aries Blueprint Core 1.4.2 Apache License Version 2.0 Apache Aries Blueprint Core 1.0.0 Apache License Version 2.0 Compatiblity Fragment Bundle Apache Aries JMX API 1.1.1 Apache License Version 2.0 Apache Aries JMX Blueprint API 1.1.0 Apache License Version 2.0 Apache Aries JMX Blueprint Core 1.1.0 Apache License Version 2.0 Apache Aries JMX Core 1.1.2 Apache License Version 2.0 Apache Aries JMX Whiteboard 1.0.0 Apache License Version 2.0 Apache Aries Proxy API 1.0.1 Apache License Version 2.0 Apache Aries Proxy Service 1.0.4 Apache License Version 2.0 Apache Aries Quiesce API 1.0.0 Apache License Version 2.0 Apache -

Technology Overview
Big Data Technology Overview Term Description See Also Big Data - the 5 Vs Everyone Must Volume, velocity and variety. And some expand the definition further to include veracity 3 Vs Know and value as well. 5 Vs of Big Data From Wikipedia, “Agile software development is a group of software development methods based on iterative and incremental development, where requirements and solutions evolve through collaboration between self-organizing, cross-functional teams. Agile The Agile Manifesto It promotes adaptive planning, evolutionary development and delivery, a time-boxed iterative approach, and encourages rapid and flexible response to change. It is a conceptual framework that promotes foreseen tight iterations throughout the development cycle.” A data serialization system. From Wikepedia, Avro Apache Avro “It is a remote procedure call and serialization framework developed within Apache's Hadoop project. It uses JSON for defining data types and protocols, and serializes data in a compact binary format.” BigInsights Enterprise Edition provides a spreadsheet-like data analysis tool to help Big Insights IBM Infosphere Biginsights organizations store, manage, and analyze big data. A scalable multi-master database with no single points of failure. Cassandra Apache Cassandra It provides scalability and high availability without compromising performance. Cloudera Inc. is an American-based software company that provides Apache Hadoop- Cloudera Cloudera based software, support and services, and training to business customers. Wikipedia - Data Science Data science The study of the generalizable extraction of knowledge from data IBM - Data Scientist Coursera Big Data Technology Overview Term Description See Also Distributed system developed at Google for interactively querying large datasets. Dremel Dremel It empowers business analysts and makes it easy for business users to access the data Google Research rather than having to rely on data engineers. -

Kafka Avro Schema Validation
Kafka Avro Schema Validation Acetous Edouard still diagnoses: unwithering and scaphocephalic Vincents elegizing quite self-denyingly but job her Elastoplasts debauchedly. Self-flattering and racy Delbert builds almost last, though Lucius excoriated his relay demagnetizes. Acidulated Torin mongrelize haltingly and prismatically, she vocalizes her sentries disannulling immanely. Here is kafka schema registry can add either json Avro is each data serialization system that serializes data then a user-specified schema The schema is closure in JSON format and describes the fields and their types Here found an. Avrors Rust Docsrs. The mad is over broker-side schema validation is girl in CP. Note follow the refrigerator of use attribute need be any relative or absolute. Senior at Wellesley College studying Media Arts and Sciences. If you sure you choose a kafka avro schema validation wtih spring. LDAP example decrease the previous section. Confluent Schema Registry stores Avro Schemas for Kafka. When a producer produces an edible, the Schema Registry is searched. If population were using Avro with Confluent's Schema Registry we'd need. Here's another example to an Avro schema in JSON from the Avro specification. This tells you what citizen data view is. AVRO is simply remote procedure call back data serialization framework designed for the bypass of defining data types and protocols and serialising data only a compact binary format. XML file as a she or CSV file. Using apache kafka log file view, validating json to json file which shows its entire content delivery. All data with kafka avro schema validation. Kafka Producer StreamSets. Avro format. -

Code Smell Prediction Employing Machine Learning Meets Emerging Java Language Constructs"
Appendix to the paper "Code smell prediction employing machine learning meets emerging Java language constructs" Hanna Grodzicka, Michał Kawa, Zofia Łakomiak, Arkadiusz Ziobrowski, Lech Madeyski (B) The Appendix includes two tables containing the dataset used in the paper "Code smell prediction employing machine learning meets emerging Java lan- guage constructs". The first table contains information about 792 projects selected for R package reproducer [Madeyski and Kitchenham(2019)]. Projects were the base dataset for cre- ating the dataset used in the study (Table I). The second table contains information about 281 projects filtered by Java version from build tool Maven (Table II) which were directly used in the paper. TABLE I: Base projects used to create the new dataset # Orgasation Project name GitHub link Commit hash Build tool Java version 1 adobe aem-core-wcm- www.github.com/adobe/ 1d1f1d70844c9e07cd694f028e87f85d926aba94 other or lack of unknown components aem-core-wcm-components 2 adobe S3Mock www.github.com/adobe/ 5aa299c2b6d0f0fd00f8d03fda560502270afb82 MAVEN 8 S3Mock 3 alexa alexa-skills- www.github.com/alexa/ bf1e9ccc50d1f3f8408f887f70197ee288fd4bd9 MAVEN 8 kit-sdk-for- alexa-skills-kit-sdk- java for-java 4 alibaba ARouter www.github.com/alibaba/ 93b328569bbdbf75e4aa87f0ecf48c69600591b2 GRADLE unknown ARouter 5 alibaba atlas www.github.com/alibaba/ e8c7b3f1ff14b2a1df64321c6992b796cae7d732 GRADLE unknown atlas 6 alibaba canal www.github.com/alibaba/ 08167c95c767fd3c9879584c0230820a8476a7a7 MAVEN 7 canal 7 alibaba cobar www.github.com/alibaba/ -

Release Notes Date Published: 2020-10-13 Date Modified
Cloudera Runtime 7.1.4 Release Notes Date published: 2020-10-13 Date modified: https://docs.cloudera.com/ Legal Notice © Cloudera Inc. 2021. All rights reserved. The documentation is and contains Cloudera proprietary information protected by copyright and other intellectual property rights. No license under copyright or any other intellectual property right is granted herein. Copyright information for Cloudera software may be found within the documentation accompanying each component in a particular release. Cloudera software includes software from various open source or other third party projects, and may be released under the Apache Software License 2.0 (“ASLv2”), the Affero General Public License version 3 (AGPLv3), or other license terms. Other software included may be released under the terms of alternative open source licenses. Please review the license and notice files accompanying the software for additional licensing information. Please visit the Cloudera software product page for more information on Cloudera software. For more information on Cloudera support services, please visit either the Support or Sales page. Feel free to contact us directly to discuss your specific needs. Cloudera reserves the right to change any products at any time, and without notice. Cloudera assumes no responsibility nor liability arising from the use of products, except as expressly agreed to in writing by Cloudera. Cloudera, Cloudera Altus, HUE, Impala, Cloudera Impala, and other Cloudera marks are registered or unregistered trademarks in the United States and other countries. All other trademarks are the property of their respective owners. Disclaimer: EXCEPT AS EXPRESSLY PROVIDED IN A WRITTEN AGREEMENT WITH CLOUDERA, CLOUDERA DOES NOT MAKE NOR GIVE ANY REPRESENTATION, WARRANTY, NOR COVENANT OF ANY KIND, WHETHER EXPRESS OR IMPLIED, IN CONNECTION WITH CLOUDERA TECHNOLOGY OR RELATED SUPPORT PROVIDED IN CONNECTION THEREWITH. -

Preview AVRO Tutorial
Avro About the Tutorial Apache Avro is a language-neutral data serialization system, developed by Doug Cutting, the father of Hadoop. This is a brief tutorial that provides an overview of how to set up Avro and how to serialize and deserialize data using Avro. Audience This tutorial is prepared for professionals aspiring to learn the basics of Big Data Analytics using Hadoop Framework and become a successful Hadoop developer. It will be a handy resource for enthusiasts who want to use Avro for data serialization and deserialization. Prerequisites Before you start proceeding with this tutorial, we assume that you are already aware of Hadoop's architecture and APIs, and you have experience in writing basic applications, preferably using Java. Disclaimer & Copyright Copyright 2015 by Tutorials Point (I) Pvt. Ltd. All the content and graphics published in this e-book are the property of Tutorials Point (I) Pvt. Ltd. The user of this e-book is prohibited to reuse, retain, copy, distribute or republish any contents or a part of contents of this e-book in any manner without written consent of the publisher. We strive to update the contents of our website and tutorials as timely and as precisely as possible, however, the contents may contain inaccuracies or errors. Tutorials Point (I) Pvt. Ltd. provides no guarantee regarding the accuracy, timeliness or completeness of our website or its contents including this tutorial. If you discover any errors on our website or in this tutorial, please notify us at [email protected]. i Avro Table of Contents About the Tutorial .........................................................................................................................................