Recruitment of Leukemic Cells from G 0 Phase of the Cell Cycle By
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Epithelial Cell Death Analysis of Cell Cycle by Flow Cytometry White Paper
Epithelial Cell Death Analysis of Cell Cycle by Flow Cytometry White Paper Authors: Savithri Balasubramanian, John Tigges, Vasilis Toxavidis, Heidi Mariani. Affiliation: Beth Israel Deaconess Medical Center Harvard Stem Cell Institute a Beckman Coulter Life Sciences: Epithelial Cell Death Analysis of Cell Cycle by Flow Cytometry PRINCIPAL OF TECHNIQUE Background: Cell cycle, or cell-division cycle, is the series of events that takes place in a cell leading to its division and duplication (replication). In cells without a nucleus (prokaryotic), cell cycle occurs via a process termed binary fission. In cells with a nucleus (eukaryotes), cell cycle can be divided in two brief periods: interphase—during which the cell grows, accumulating nutrients needed for mitosis and duplicating its DNA—and the mitosis (M) phase, during which the cell splits itself into two distinct cells, often called «daughter cells». Cell-division cycle is a vital process by which a single-celled fertilized egg develops into a mature organism, as well as the process by which hair, skin, blood cells, and some internal organs are renewed. Cell cycle consists of four distinct phases: G1 phase, S phase (synthesis), G2 phase (collectively known as interphase) and M phase (mitosis). M phase is itself composed of two tightly coupled processes: mitosis, in which the cell’s chromosomes are divided between the two daughter cells, and cytokinesis, in which the cell’s cytoplasm divides in half forming distinct cells. Activation of each phase is dependent on the proper progression and completion of the previous one. Cells that have temporarily or reversibly stopped dividing are said to have entered a state of quiescence called G0 phase. -

P53 Phosphorylation and Association with Murine Double Minute 2, C-Jun
[CANCER RESEARCH 60, 896–900, February 15, 2000] Advances in Brief p53 Phosphorylation and Association with Murine Double Minute 2, c-Jun ARF NH2-Terminal Kinase, p14 , and p300/CBP during the Cell Cycle and after Exposure to Ultraviolet Irradiation1 Thomas Buschmann, Victor Adler, Ekaterina Matusevich, Serge Y. Fuchs, and Ze’ev Ronai2 Ruttenberg Cancer Center, Mount Sinai School of Medicine, New York, New York 10029 Abstract though the mechanisms underlying the ability of p53 to elicit such opposing effects are yet to be identified, independent studies point to p53 phosphorylation and association with proteins is implicated in its a different set of p53 regulators and effectors that are affected by p53 stability and activity. We have compared the association of DNA-bound in each of these scenarios. and overall pools of p53 with murine double minute 2 (Mdm2), c-Jun NH -terminal kinase (JNK), p300/CBP, and p14ARF during cell cycle In studying the regulation of p53 stability, we demonstrated previ- 2 ously that, in Swiss 3T3 cells, JNK and Mdm2 target p53 degradation progression. Whereas DNA-bound p53 associates with JNK at G0-G1 and in different phases of the cell cycle (7). In this study, we have with Mdm2 and p300 during S and G2-M phases, the general pool of p53 was found in complex with JNK and Mdm2 almost throughout the cell compared the association of subpopulations of p53 with proteins cycle. Phosphorylation of p53 at serines 9, 15, and 20 is at the highest levels implicated in stability and activity of p53 and monitored the pattern of at G1 and at serines 37 and 392 during G2-M phase. -

A P53-Responsive Mirna Network Promotes Cancer Cell Quiescence Ting La1, Guang Zhi Liu2, Margaret Farrelly1, Nicole Cole3, Yu Chen Feng1, Yuan Yuan Zhang1, Simonne K
Published OnlineFirst October 9, 2018; DOI: 10.1158/0008-5472.CAN-18-1886 Cancer Tumor Biology and Immunology Research A p53-Responsive miRNA Network Promotes Cancer Cell Quiescence Ting La1, Guang Zhi Liu2, Margaret Farrelly1, Nicole Cole3, Yu Chen Feng1, Yuan Yuan Zhang1, Simonne K. Sherwin1, Hamed Yari1, Hessam Tabatabaee1, Xu Guang Yan1, Su Tang Guo4, Tao Liu5, Rick F. Thorne2,6, Lei Jin7, and Xu Dong Zhang1,2 Abstract Cancer cells in quiescence (G0 phase) are resistant to miRNA-455-3p targeted CDK2-associated cullin domain 1 death, and re-entry of quiescent cancer cells into the cell- (CAC1), which enhanced CDK2-mediated phosphorylation cycle plays an important role in cancer recurrence. Here we of p27 necessary for its polyubiquitination. Of note, the show that two p53-responsive miRNAs utilize distinct but gene encoding miRNA-27b-3p was embedded in the intron complementary mechanisms to promote cancer cell quies- of the chromosome 9 open reading frame 3 gene that was cence by facilitating stabilization of p27. Purified quiescent transcriptionally activated by p53. Similarly, the host gene B16 mouse melanoma cells expressed higher levels of of miRNA-455-3p, collagen alpha-1 (XXVII) chain, was also a miRNA-27b-3p and miRNA-455-3p relative to their prolif- p53 transcriptional target. Collectively, our results identify erating counterparts. Induction of quiescence resulted in miRNA-27b-3p and miRNA-455-3p as important regulators increased levels of these miRNAs in diverse types of human of cancer cell quiescence in response to p53 and suggest that cancer cell lines. Inhibition of miRNA-27b-3p or miRNA- manipulating miRNA-27b-3p and miRNA-455-3p may con- 455-3p reduced, whereas its overexpression increased, the stitute novel therapeutic avenues for improving outcomes of proportion of quiescent cells in the population, indicating cancer treatment. -

The Cyclin-Dependent Kinase Inhibitor and Tumor Suppressor Locus P16/INK4A- P14arf and Regulation of the Transition Into and out of the Cell Cycle
The Cyclin-Dependent Kinase Inhibitor and Tumor Suppressor Locus p16/INK4A- p14ARF and Regulation of the Transition Into and Out of the Cell Cycle by Payal Agarwal A dissertation submitted to the Graduate Faculty of Auburn University in partial fulfillment of the requirements for the Degree of Doctor of Philosophy Auburn, Alabama May 7, 2012 Copyright, 2011 by Payal Agarwal Approved by Richard C. Bird, Chair, Professor of Pathobiology Bruce F. Smith, Professor of Pathobiology Frederik W. van Ginkel, Associate Professor of Pathobiology Anthony G. Moss, Associate Professor of Biological Sciences Abstract p16/INK4A/CDKN2A is an important tumor suppressor gene located in the INK4A/ARF locus, which encodes a 16 kDa protein known as p16, and a 14 kDa protein known as p14ARF in humans. p16 arrests cell cycle in early G1 phase thereby inhibiting the binding of cyclin dependent kinase 4/6 with cyclinD1. This leaves the retinoblastoma protein (pRb) tumor suppressor hypo-phosphorylated and S phase transcription factor E2F bound and inactive. p14ARF expression up-regulates cyclin dependent kinase inhibitor p21, which inhibits the G1/S phase transition by stabilizing p53 expression upon disassociation from mdm2. We hypothesized that p16 has a role in exit from the cell cycle, becomes defective in cancer cells and has binding partners other than CDK4/CDK6 in quiescent or differentiated cells when their canonical target proteins are thought to be nonfunctional. We have hypothesized that INK4A/ARF encoded proteins perform important regulatory roles that are defective in canine mammary cancer and may cause loss of differentiation potential. Well characterized p16-defective canine mammary cancer cell lines, normal canine fibroblasts, and CMT-derived p16-transfected CMT cell clones, are used to investigate expression of p16 after serum starvation into quiescence followed by re-feeding to induce cell cycle re-entry. -

Exit from Quiescence Displays a Memory of Cell Growth and Division
ARTICLE DOI: 10.1038/s41467-017-00367-0 OPEN Exit from quiescence displays a memory of cell growth and division Xia Wang1,2, Kotaro Fujimaki1, Geoffrey C. Mitchell1,3, Jungeun Sarah Kwon1, Kimiko Della Croce1, Chris Langsdorf4, Hao Helen Zhang5 & Guang Yao1,6 Reactivating quiescent cells to proliferate is critical to tissue repair and homoeostasis. Quiescence exit is highly noisy even for genetically identical cells under the same environmental conditions. Deregulation of quiescence exit is associated with many diseases, but cellular mechanisms underlying the noisy process of exiting quiescence are poorly understood. Here we show that the heterogeneity of quiescence exit reflects a memory of preceding cell growth at quiescence induction and immediate division history before quiescence entry, and that such a memory is reflected in cell size at a coarse scale. The deterministic memory effects of preceding cell cycle, coupled with the stochastic dynamics of an Rb-E2F bistable switch, jointly and quantitatively explain quiescence-exit heterogeneity. As such, quiescence can be defined as a distinct state outside of the cell cycle while displaying a sequential cell order reflecting preceding cell growth and division variations. 1 Department of Molecular and Cellular Biology, University of Arizona, Tucson, AZ 85721, USA. 2 School of Biological and Medical Engineering, Hefei University of Technology, Hefei, Anhui 230009, China. 3 Department of Biology, Wofford College, Spartanburg, SC 29303, USA. 4 Molecular Probes, Thermo Fisher Scientific, Eugene, OR 97402, USA. 5 Department of Mathematics, University of Arizona, Tucson, AZ 85721, USA. 6 Arizona Cancer Centre, University of Arizona, Tucson, AZ 85719, USA. Xia Wang, Kotaro Fujimaki and Geoffrey C. -

Detailed Description of Gene Signatures
Detailed description of gene signatures Immune response gene signatures Four QTC clusters showed a significant association with immune response, QTC1, QTC8, QTC10, and QTC17. QTC1 was highly enriched for GO:0002376 immune system process (p=9.2x10-31) and GO:0046649 lymphocyte activation (7.1x10-7), QTC8 for GO:0002376 immune system processes (p=9.7x10-5), QTC10 for GO:0002474 antigen processing and presentation of peptide antigen via MHC class I (p=2.1x10-5), and QTC17 for the KEGG pathway hsa04060 cytokine-cytokine receptor interaction (p=2.3x10-5), suggesting that these gene clusters represents different aspects of the immune response system. QTC1 and QTC8 were highly correlated (r=0.85) whereas QTC10 and QTC17 appeared more independent. QTC1 and QTC8 were more strongly expressed in the MS2b.1 subtype whereas QTC10 and QTC17 were more strongly expressed in the MS2b2.2 subtype QTC1 and QTC8 were merged and subsets of immune-related genes are shown in Figures 1A and 1B. The signature had a prominent activated T-cell theme including key genes ranging from T-cell stimulatory chemokines, T-cell receptor complex genes, T-cell specific markers, as well as signal transducers and effector genes of cytotoxic T-cells. Among the early T-cell activator genes were CCL2, CCL5, IL2RA, IL2RB, and IL7R. CCL2 and CCL5 are known to be produced by tumor cells and induce transendothelial migration and chemotaxis of effector T-cells (Brown et al. 2007, Soria and Ben-Baruch, 2008). CCL5 co-localizes with tumor infiltrating leukocytes and the CCL5 concentrations correlates to CD8+ T-cell infiltration in the epithelial areas of tumors (Negus et al. -

Cell Cycle Wikipedia Cell Cycle from Wikipedia, the Free Encyclopedia
11/8/2016 Cell cycle Wikipedia Cell cycle From Wikipedia, the free encyclopedia The cell cycle or celldivision cycle is the series of events that take place in a cell leading to its division and duplication of its DNA (DNA replication) to produce two daughter cells. In bacteria, which lack a cell nucleus, the cell cycle is divided into the B, C, and D periods. The B period extends from the end of cell division to the beginning of DNA replication. DNA replication occurs during the C period. The D period refers to the stage between the end of DNA replication and the splitting of the bacterial cell into two daughter cells.[2] In cells with a nucleus, as in eukaryotes, the cell cycle is also divided into three periods: interphase, the mitotic (M) phase, and cytokinesis. During interphase, the cell grows, accumulating nutrients needed for mitosis, preparing it for cell division and duplicating its DNA. During the mitotic phase, the chromosomes separate. During the final stage, cytokinesis, the chromosomes and cytoplasm separate into two new daughter cells. To ensure the proper division of the cell, there are control mechanisms known as cell cycle checkpoints. Life Cycle of the cell The celldivision cycle is a vital process by which a singlecelled fertilized egg develops into a mature organism, as well as the process by which hair, skin, blood cells, and some internal organs are renewed. After cell division, each of the daughter cells begin the interphase of a new cycle. Although the various stages of interphase are not usually morphologically distinguishable, each phase of the cell cycle has a distinct set of specialized biochemical processes that prepare the cell for initiation of cell division. -

ABT-263 Induces G1/G0-Phase Arrest, Apoptosis and Autophagy in Human Esophageal Cancer Cells in Vitro
Acta Pharmacologica Sinica 2017: 1632–1641 © 2017 CPS and SIMM All rights reserved 1671-4083/17 www.nature.com/aps Original Article ABT-263 induces G1/G0-phase arrest, apoptosis and autophagy in human esophageal cancer cells in vitro Qing-huan LIN, Fu-chang QUE, Chun-ping GU, De-sheng ZHONG, Dan ZHOU, Yi KONG, Le YU*, Shu-wen LIU* State Key Laboratory of Organ Failure Research, Guangdong Provincial Key Laboratory of New Drug Screening, School of Pharmaceutical Sciences, Southern Medical University, Guangzhou 510515, China Abstract Both the anti- and pro-apoptotic members of the Bcl-2 family are regulated by a conserved Bcl-2 homology (BH3) domain. ABT- 263 (Navitoclax), a novel BH3 mimetic and orally bioavailable Bcl-2 family inhibitor with high affinity for Bcl-xL, Bcl-2 and Bcl-w has entered clinical trials for cancer treatment. But the anticancer mechanisms of ABT-263 have not been fully elucidated. In this study we investigated the effects of ABT-263 on human esophageal cancer cells in vitro and to explore its anticancer mechanisms. Treatment with ABT-263 dose-dependently suppressed the viability of 3 human esophageal cancer cells with IC50 values of 10.7±1.4, 7.1±1.5 and 8.2±1.6 μmol/L, in EC109, HKESC-2 and CaES-17 cells, respectively. ABT-263 (5–20 μmol/L) dose-dependently induced G1/G0-phase arrest in the 3 cancer cell lines and induced apoptosis evidenced by increased the Annexin V-positive cell population and elevated levels of cleaved caspase 3, cleaved caspase 9 and PARP. -
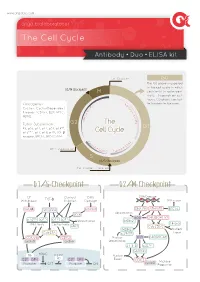
The Cell Cycle
www.arigobio.com The Cell Cycle Antibody˙Duo˙ELISA kit Cell Division G0 The G0 phase is a period in the cell cycle in which G2/M Checkpoint M cells exist in quiescent state. Depends on cell types, G0 phase can last 6 C / for few days to few years. y Oncogenes : 4 c K l i n Cyclins, Cyclin-Dependent D B C + + Kinases (CDKs), E2F, MYC, C D d MDM2. n i c l 2 c y Tumor Suppressors : G2 The C RB, p53, p21, p27, p57, p14ARF, G1 p16INK4a, p17, p18, p19, TGF-β Cell Cycle receptor, BRCA1, BRCA2, ATM. C y 2 c K lin D A C + + DNA Duplication CD inE K1/2 Cycl S G1/S Checkpoint Cell Content Doubled G1/S Checkpoint G2/M Checkpoint GF Contact DNA DNA Damage Withdrawal TGF-β Innibition Damage DNA repair GSK3β Smad3/4 ATM/ATR DNA-PK ATM/ATR BRCA1 p53 Ubiquitination INK4 p53 CHK2 CHK1/2 p16 p15 p27 p21 Ubiquitination MDM2 INK4 family Cip/Kip family 14-3-3 SKP2 Cdc25B/C MDM2 Nuclear Cdc25A p19ARF Export CDK4/6 CDK2 Nuclear p53 p300/PCAF cyclinD cyclinE Sequestration 14-3-3 p21Cip1 GADD45 Rb HDAC Nuclear Rb E2F DP-I E2F DP-I Export CDC2 cyclinB M phase Promoter OFF Promoter ON Progression Common Methods for Mammalian Cell Cycle Synchronization Cell Cycle Stage Blocked Method Proposed mechanism of action G0/G1 Serum Starvation Growth restriction Lovastin Inhibition of HMG-CoA reductase G1 Mimosine Inhibit Thymidine, nucleotide biosynthesis Thymidine Feedback inhibition of DNA replication G1/S Aphidicolin Inhibition of DNA polymerases Hydroxyurea Inhibition of ribonucleotide reductase Colchicine/Colcemide Inhibition of microtubule polymerization G2/M Nocodazole Inhibition of microtubule polymerization ARG30139 Cell Cycle Marker Antibody Panel (Cyclin B1, Cyclin E) The cell cycle is controlled by numerous mechanisms ensuring correct cell division. -

The Roles of Cyclin-Dependent Kinases in Cell-Cycle Progression and Therapeutic Strategies in Human Breast Cancer
International Journal of Molecular Sciences Review The Roles of Cyclin-Dependent Kinases in Cell-Cycle Progression and Therapeutic Strategies in Human Breast Cancer 1,2, 1,2, 1,2, 1,2 1,2 Lei Ding y, Jiaqi Cao y, Wen Lin y, Hongjian Chen , Xianhui Xiong , Hongshun Ao 1,2, Min Yu 1,2, Jie Lin 1,2 and Qinghua Cui 1,2,* 1 Lab of Biochemistry & Molecular Biology, School of Life Sciences, Yunnan University, Kunming 650091, China; [email protected] (L.D.); [email protected] (J.C.); [email protected] (W.L.); [email protected] (H.C.); [email protected] (X.X.); [email protected] (H.A.); [email protected] (M.Y.); [email protected] (J.L.) 2 Key Lab of Molecular Cancer Biology, Yunnan Education Department, Kunming 650091, China * Correspondence: [email protected] The authors contributed equally to this work. y Received: 31 December 2019; Accepted: 24 February 2020; Published: 13 March 2020 Abstract: Cyclin-dependent kinases (CDKs) are serine/threonine kinases whose catalytic activities are regulated by interactions with cyclins and CDK inhibitors (CKIs). CDKs are key regulatory enzymes involved in cell proliferation through regulating cell-cycle checkpoints and transcriptional events in response to extracellular and intracellular signals. Not surprisingly, the dysregulation of CDKs is a hallmark of cancers, and inhibition of specific members is considered an attractive target in cancer therapy. In breast cancer (BC), dual CDK4/6 inhibitors, palbociclib, ribociclib, and abemaciclib, combined with other agents, were approved by the Food and Drug Administration (FDA) recently for the treatment of hormone receptor positive (HR+) advanced or metastatic breast cancer (A/MBC), as well as other sub-types of breast cancer. -

Infectious Delivery of the 132 Kb CDKN2A/CDKN2B Genomic DNA Region Results in Correctly Spliced Gene Expression and Growth Suppression in Glioma Cells
Gene Therapy (2004) 11, 1195–1204 & 2004 Nature Publishing Group All rights reserved 0969-7128/04 $30.00 www.nature.com/gt RESEARCH ARTICLE Infectious delivery of the 132 kb CDKN2A/CDKN2B genomic DNA region results in correctly spliced gene expression and growth suppression in glioma cells R Inoue, KA Moghaddam, M Ranasinghe, Y Saeki, EA Chiocca and R Wade-Martins1 Molecular Neuro-Oncology Laboratories, Neurosurgery Service, Massachusetts General Hospital-East and Harvard Medical School, Charlestown, MA, USA The expression of genes from genomic loci can be relatively these proteins are expressed from overlapping genes, complex, utilizing exonic, intronic and flanking sequences utilizing alternative splicing and promoter usage. The to regulate tissue and developmental specificity. Infectious delivered locus expresses each gene at physiological levels bacterial artificial chromosomes (iBACs) have been shown to and cellular responses (apoptosis versus growth arrest) deliver and express large genomic loci (up to 135 kb) into occur dependent on cellular p53 status, as expected. The primary cells for functional analyses. The delivery of large work further demonstrates the potential of the iBAC system genomic DNA inserts allows the expression of complex loci for the delivery of genomic loci whose expression is mediated and of multiple splice variants. Herein, we demonstrate for by complex splicing and promoter usage both for gene the first time that an iBAC will deliver and correctly express in therapy applications and functional genomics studies. human glioma cells the entire CDKN2A/CDKN2B genomic Gene Therapy (2004) 11, 1195–1204. doi:10.1038/ region, which encodes for at least three important cell-cycle sj.gt.3302284; Published online 27 May 2004 regulatory proteins (p16INK4a, p14ARF and p15INK4b). -

Spindle Checkpoint Function Is Required for Mitotic Catastrophe Induced by DNA-Damaging Agents
Oncogene (2004) 23, 6548–6558 & 2004 Nature Publishing Group All rights reserved 0950-9232/04 $30.00 www.nature.com/onc Spindle checkpoint function is required for mitotic catastrophe induced by DNA-damaging agents Masayuki Nitta1,2,3, Osamu Kobayashi1,2,3, Shinobu Honda1, Toru Hirota1, Shinji Kuninaka1, Tomotoshi Marumoto1, Yukitaka Ushio2 and Hideyuki Saya*,1 1Department of Tumor Genetics and Biology, Graduate School of Medical Sciences, University of Kumamoto, 1-1-1 Honjo, Kumamoto 860-8556, Japan; 2Department of Neurosurgery, Graduate School of Medical Sciences, University of Kumamoto, Kumamoto, Japan Mitotic catastrophe is an important mechanism for the ments such as chemotherapy and radiation. Most cancer induction of cell death in cancer cells by antineoplastic cells are deficient in G1 checkpoint function and agents that damage DNA. This process is facilitated by therefore fail to arrest in G1 phase on exposure to geno- defects in the G1 and G2 checkpoints of the cell cycle that toxic agents. Instead, they accumulate temporarily in are apparent in most cancer cells and which allow the cells G2 phase. However, given that the G2 checkpoint is to enter mitosis with DNA damage. We have now also partially impaired in cancer cells, they are unable characterized the dynamics of mitotic catastrophe induced to maintain G2 arrest and eventually die as they enter by DNA-damaging agents in p53-deficient cancer cells. mitosis (Fingert et al., 1986; Chan et al., 1999). This Cells that entered mitosis with DNA damage transiently process is known as mitotic catastrophe or mitotic death arrested at metaphase for more than 10 h without (Chan et al., 1999; Roninson et al., 2001).