Usage and Demo of R Package Ncov2019
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Research Article Stability and Complexity Analysis of Temperature Index Model Considering Stochastic Perturbation
Hindawi Advances in Mathematical Physics Volume 2018, Article ID 2789412, 18 pages https://doi.org/10.1155/2018/2789412 Research Article Stability and Complexity Analysis of Temperature Index Model Considering Stochastic Perturbation Jing Wang Faculty of Science, Bengbu University, Bengbu 233030, China Correspondence should be addressed to Jing Wang; [email protected] Received 20 October 2017; Accepted 12 December 2017; Published 1 January 2018 Academic Editor: Giampaolo Cristadoro Copyright © 2018 Jing Wang. Tis is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. A temperature index model with delay and stochastic perturbation is constructed in this paper. It explores the infuence of parameters and stochastic factors on the stability and complexity of the model. Based on historical temperature data of four cities of Anhui Province in China, the temperature periodic variation trends of approximately sinusoidal curves of four cities are given, respectively. In addition, we analyze the existence conditions of the local stability of the temperature index model without stochastic term and estimate its parameters by using the same historical data of the four cities, respectively. Te numerical simulation results of the four cities are basically consistent with the descriptions of their historical temperature data, which proves that the temperature index model constructed has good ftting degree. It also shows that unreasonable delay parameter can make the model lose stability and improve the complexity. Stochastic factors do not usually change the trend in temperature, but they can cause high frequency fuctuations in the process of temperature evolution. -

Analysis of Historic Rainfall Characteristics for Robust Wheat Cropping in North Anhui
Analysis of historic rainfall characteristics for robust wheat cropping in North Anhui Hui Su, Yibo Wu, Yulei Zhu, Youhong Song Anhui Agricultural University, School of Agronomy, Hefei, 233036. Corresponding email: [email protected] Abstract Northern part of Anhui is one of major wheat producing areas in China. The total amount of rainfall is sufficient for wheat season; however, it is unevenly distributed at the different growth stages, resulting in risk of yield losses. In order to optimise the cultivation in North Anhui, it is essential to characterise the rainfall pattern for wheat growth particularly in the critical period (i.e. the months of sowing and harvesting). By analysing the rainfall data from 1955 to 2017, this study characterised the rainfall pattern from six sites representing different regions of North Anhui. The frequency of continuous rainfall days during sowing and harvesting periods were quantified based on 63 years rainfall distribution. The characterisation of rainfall in six representative sites in North Anhui were able to be used to guide wheat sowing and harvesting, which could help farmers to make decisions and avoid likelihood of cropping risks. Key Words Winter wheat, rainfall pattern, sustainable cropping, China Introduction Huang-Huai-Hai plain is a major wheat production area in China (Yang, 2018). The northern part of Anhui province belongs to south Huang-Huai-Hai plain. The wheat sowing time normally varies 1 week before or after October 15 and harvesting is tightly closing to May 31 in the following year. The climate characteristics particularly rainfall distribution over the season are complicated in North Anhui as it is in the transitional zone between the North and South China. -

Study on the Spatial-Temporal Pattern Evolution and Countermeasures of Regional Coordinated Development in Anhui Province, China
Current Urban Studies, 2020, 8, 115-128 https://www.scirp.org/journal/cus ISSN Online: 2328-4919 ISSN Print: 2328-4900 Study on the Spatial-Temporal Pattern Evolution and Countermeasures of Regional Coordinated Development in Anhui Province, China Yizhen Zhang1,2*, Weidong Cao1,2, Kun Zhang3 1School of Geography and Tourism, Anhui Normal University, Wuhu, China 2Urban and Regional Planning Research Center of Anhui Normal University, Wuhu, China 3College of Tourism, Huaqiao University, Quanzhou, China How to cite this paper: Zhang, Y. Z., Cao, Abstract W. D., & Zhang, K. (2020). Study on the Spatial-Temporal Pattern Evolution and Regional coordinated development was an important measure to resolve new Countermeasures of Regional Coordinated contradictions in the new era, and it has gradually become a hotspot in geog- Development in Anhui Province, China. raphy research. At three time points of 2010, 2013, and 2017, by constructing Current Urban Studies, 8, 115-128. an evaluation index system for the city comprehensive competitiveness, the https://doi.org/10.4236/cus.2020.81005 entropy method and the coupling coordination model were used to study the Received: March 2, 2020 coordinated development pattern of cities in Anhui Province. Moreover, we Accepted: March 27, 2020 tried to raise the issue of regional coordinated development in Anhui Prov- Published: March 30, 2020 ince and corresponding countermeasures. The results showed that the com- Copyright © 2020 by author(s) and prehensive competitiveness of cities in Anhui Province has strong spatio- Scientific Research Publishing Inc. temporal heterogeneity. The spatial development pattern with Hefei as the This work is licensed under the Creative core was more obvious. -

Quantitative Analysis of Burden of Infectious Diarrhea Associated with Floods in Northwest of Anhui Province, China: a Mixed Method Evaluation
Quantitative Analysis of Burden of Infectious Diarrhea Associated with Floods in Northwest of Anhui Province, China: A Mixed Method Evaluation Guoyong Ding1, Ying Zhang2, Lu Gao1, Wei Ma1, Xiujun Li1, Jing Liu1, Qiyong Liu3, Baofa Jiang1* 1 Department of Epidemiology and Health Statistics, School of Public Health, Shandong University, Jinan City, Shandong Province, P.R. China, 2 School of Public Health, China Studies Centre, The University of Sydney, New South Wales, Australia, 3 National Institute for Communicable Disease Control and Prevention, China CDC, Beijing City, P.R. China Abstract Background: Persistent and heavy rainfall in the upper and middle Huaihe River of China brought about severe floods during the end of June and July 2007. However, there has been no assessment on the association between the floods and infectious diarrhea. This study aimed to quantify the impact of the floods in 2007 on the burden of disease due to infectious diarrhea in northwest of Anhui Province. Methods: A time-stratified case-crossover analysis was firstly conducted to examine the relationship between daily cases of infectious diarrhea and the 2007 floods in Fuyang and Bozhou of Anhui Province. Odds ratios (ORs) of the flood risk were quantified by conditional logistic regression. The years lived with disability (YLDs) of infectious diarrhea attributable to floods were then estimated based on the WHO framework of the calculating potential impact fraction in the Burden of Disease study. Results: A total of 197 infectious diarrheas were notified during the exposure and control periods in the two study areas. The strongest effect was shown with a 2-day lag in Fuyang and a 5-day lag in Bozhou. -

Quantitative Assessment and Diagnosis for Regional Agricultural Drought Resilience Based on Set Pair Analysis and Connection Entropy
Article Quantitative Assessment and Diagnosis for Regional Agricultural Drought Resilience Based on Set Pair Analysis and Connection Entropy Menglu Chen 1, Shaowei Ning 1, Yi Cui 2,*, Juliang Jin 1, Yuliang Zhou 1 and Chengguo Wu 1 1 School of Civil Engineering, Hefei University of Technology, Hefei 230009, China; [email protected] (M.C.); [email protected] (S.N.); [email protected] (J.J.); [email protected] (Y.Z.); [email protected] (C.W.) 2 State Key Laboratory of Hydraulic Engineering Simulation and Safety, Tianjin University, Tianjin 300072, China * Correspondence: [email protected]; Tel.: +86-13102218320 Received: 27 February 2019; Accepted: 02 April 2019; Published: 5 April 2019 Abstract:Assessment and diagnosis of regional agricultural drought resilience (RADR) is an important groundwork to identify the shortcomings of regional agriculture to resist drought disasters accurately. In order to quantitatively assess the capacity of regional agriculture system to reduce losses from drought disasters under complex conditions and to identify vulnerability indexes, an assessment and diagnosis model for RADR was established. Firstly, this model used the improved fuzzy analytic hierarchy process to determine the index weights, then proposed an assessment method based on connection number and an improved connection entropy. Furthermore, the set pair potential based on subtraction was used to diagnose the vulnerability indexes. In addition, a practical application had been carried out in the region of the Huaibei Plain in Anhui Province. The evaluation results showed that the RADR in this area from 2005 to 2014 as a whole was in a relatively weak situation. However, the average grade values had decreased from 3.144 to 2.790 during these 10 years and the RADR had an enhanced tendency. -

PRC: Anhui Integrated Transport Sector Improvement Project–Xuming Expressway
Updated Resettlement Plan January 2011 PRC: Anhui Integrated Transport Sector Improvement Project–Xuming Expressway Prepared by Anhui Provincial Communications Investment Group Company for the Asian Development Bank. 2 CURRENCY EQUIVALENTS Currency unit – Yuan (CNY) $1.00 = CNY6.80 ABBREVIATIONS ACTVC - Anhui Communications Vocational & Technical College ADB - Asian Development Bank AHAB - Anhui Highway Administration Bureau APCD - Anhui Provincial Communications Department Anhui Provincial Communications Investment Group ACIG - Company APs Affected Persons AVs Affected Villages APG - Anhui Provincial Government M&E Monitoring and Evaluation PMO - Project Management Office RP - Resettlement Plan PRC - People’s Republic of China NOTE (i) In this report, "$" refers to US dollars unless otherwise stated. This updated resettlement plan is a document of the borrower. The views expressed herein do not necessarily represent those of ADB's Board of Directors, Management, or staff, and may be preliminary in nature. Your attention is directed to the “terms of use” section of this website. In preparing any country program or strategy, financing any project, or by making any designation of or reference to a particular territory or geographic area in this document, the Asian Development Bank does not intend to make any judgments as to the legal or other status of any territory or area. ADB Financed Anhui Integrated Transport Sector Improvement Project Resettlement Plan for Anhui Xuzhou-Mingguang Expressway Project (updated) Anhui Provincial Communications Investment Group Company January, 2011 Note on the updated RP On November 11, 2009, the Anhui Provincial Development and Reform Commission gave a reply on the detailed design of the Anhui section of the Xuzhou-Mingguang Expressway with Document APDRC [2009] No.1199. -
Three Kingdoms Unveiling the Story: List of Works
Celebrating the 40th Anniversary of the Japan-China Cultural Exchange Agreement List of Works Organizers: Tokyo National Museum, Art Exhibitions China, NHK, NHK Promotions Inc., The Asahi Shimbun With the Support of: the Ministry of Foreign Affairs of Japan, NATIONAL CULTURAL HERITAGE ADMINISTRATION, July 9 – September 16, 2019 Embassy of the People’s Republic of China in Japan With the Sponsorship of: Heiseikan, Tokyo National Museum Dai Nippon Printing Co., Ltd., Notes Mitsui Sumitomo Insurance Co.,Ltd., MITSUI & CO., LTD. ・Exhibition numbers correspond to the catalogue entry numbers. However, the order of the artworks in the exhibition may not necessarily be the same. With the cooperation of: ・Designation is indicated by a symbol ☆ for Chinese First Grade Cultural Relic. IIDA CITY KAWAMOTO KIHACHIRO PUPPET MUSEUM, ・Works are on view throughout the exhibition period. KOEI TECMO GAMES CO., LTD., ・ Exhibition lineup may change as circumstances require. Missing numbers refer to works that have been pulled from the JAPAN AIRLINES, exhibition. HIKARI Production LTD. No. Designation Title Excavation year / Location or Artist, etc. Period and date of production Ownership Prologue: Legends of the Three Kingdoms Period 1 Guan Yu Ming dynasty, 15th–16th century Xinxiang Museum Zhuge Liang Emerges From the 2 Ming dynasty, 15th century Shanghai Museum Mountains to Serve 3 Narrative Figure Painting By Qiu Ying Ming dynasty, 16th century Shanghai Museum 4 Former Ode on the Red Cliffs By Zhang Ruitu Ming dynasty, dated 1626 Tianjin Museum Illustrated -

Research on Logistics Efficiency of Anhui Province
2019 4th International Workshop on Materials Engineering and Computer Sciences (IWMECS 2019) Research on Logistics Efficiency of Anhui Province Han Yanhui School of Traffic and Transportation, Beijing Jiaotong University, P. R. China [email protected] Keywords: Logistics efficiency; DEA model; Anhui province Abstract: China's logistics industry has developed rapidly in recent years, and logistics efficiency is the main indicator that reflects the level of logistics development. This article first illustrated the situation of logistics development in Anhui Province and established an index system for evaluation of logistics efficiency, including four input indicators, the number of employees in the logistics industry, the mileage of graded roads, investment in fixed assets in the logistics industry and education expenditure, as well as two output indicators, the freight turnover volume and the added value of the logistics industry; then use the CCR-DEA and BCC-DEA models to evaluate the logistics efficiency of 16 cities of Anhui province. The results show that there is a phenomenon of unbalanced logistics development. Among them, Suzhou, Chizhou, and Anqing have low logistics efficiency, which is the bottleneck that limits the improvement of logistics efficiency in Anhui Province. 1. Introduction With the growth of China's economic level and the continuous transformation of economic growth pattern, the development level of logistics industry has become an important index to measure the comprehensive development level of a region. Anhui province is located in the middle and lower reaches of the Yangtze river and the Huaihe river. Together with Jiangsu, Shanghai and Zhejiang, forms the Yangtze river delta urban agglomeration. -
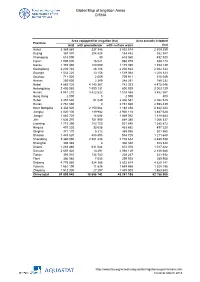
Global Map of Irrigation Areas CHINA
Global Map of Irrigation Areas CHINA Area equipped for irrigation (ha) Area actually irrigated Province total with groundwater with surface water (ha) Anhui 3 369 860 337 346 3 032 514 2 309 259 Beijing 367 870 204 428 163 442 352 387 Chongqing 618 090 30 618 060 432 520 Fujian 1 005 000 16 021 988 979 938 174 Gansu 1 355 480 180 090 1 175 390 1 153 139 Guangdong 2 230 740 28 106 2 202 634 2 042 344 Guangxi 1 532 220 13 156 1 519 064 1 208 323 Guizhou 711 920 2 009 709 911 515 049 Hainan 250 600 2 349 248 251 189 232 Hebei 4 885 720 4 143 367 742 353 4 475 046 Heilongjiang 2 400 060 1 599 131 800 929 2 003 129 Henan 4 941 210 3 422 622 1 518 588 3 862 567 Hong Kong 2 000 0 2 000 800 Hubei 2 457 630 51 049 2 406 581 2 082 525 Hunan 2 761 660 0 2 761 660 2 598 439 Inner Mongolia 3 332 520 2 150 064 1 182 456 2 842 223 Jiangsu 4 020 100 119 982 3 900 118 3 487 628 Jiangxi 1 883 720 14 688 1 869 032 1 818 684 Jilin 1 636 370 751 990 884 380 1 066 337 Liaoning 1 715 390 783 750 931 640 1 385 872 Ningxia 497 220 33 538 463 682 497 220 Qinghai 371 170 5 212 365 958 301 560 Shaanxi 1 443 620 488 895 954 725 1 211 648 Shandong 5 360 090 2 581 448 2 778 642 4 485 538 Shanghai 308 340 0 308 340 308 340 Shanxi 1 283 460 611 084 672 376 1 017 422 Sichuan 2 607 420 13 291 2 594 129 2 140 680 Tianjin 393 010 134 743 258 267 321 932 Tibet 306 980 7 055 299 925 289 908 Xinjiang 4 776 980 924 366 3 852 614 4 629 141 Yunnan 1 561 190 11 635 1 549 555 1 328 186 Zhejiang 1 512 300 27 297 1 485 003 1 463 653 China total 61 899 940 18 658 742 43 241 198 52 -

ANHUI EXPRESSWAY COMPANY LIMITED App.1A.1 (A Joint Stock Limited Company Incorporated in the People’S Republic of China with Limited Liability) App.1A 5
IMPORTANT If you are in any doubt about this prospectus, you should consult your stockbroker, bank manager, solicitor, professional accountant or other professional S38(1A) adviser. A copy of this prospectus, having attached thereto the documents specified in the section headed “Documents delivered to the Registrar of Companies in Hong Kong” in appendix X, has been registered by the Registrar of Companies in Hong Kong as required by section 342C of the Companies Ordinance of Hong S342C Kong. The Registrar of Companies in Hong Kong takes no responsibility for the contents of this prospectus or any of the documents referred to above. The Stock Exchange of Hong Kong Limited (the “Stock Exchange”) and Hong Kong Securities Clearing Company Limited (“Hongkong Clearing”) take no responsibility for the contents of this prospectus, make no representation as to its accuracy or completeness and expressly disclaim any liability whatsoever for any loss howsoever arising from or in reliance upon the whole or any part of the contents of this prospectus. ANHUI EXPRESSWAY COMPANY LIMITED App.1A.1 (a joint stock limited company incorporated in the People’s Republic of China with limited liability) App.1A 5 Placing and New Issue of 493,010,000 H Shares of nominal value RMB1.00 each, App.1A at an issue price of RMB1.89 per H Share 15(2)(a)(c)(d) payableinfullonapplication 3rd Sch at HK$1.77 per H Share App.1Apara 9 15(2)(h) Sponsors and Lead Underwriters CEF Capital Limited The CEF Group Crosby Capital Markets (Asia) Limited Co-Underwriters DBS Asia Capital Limited Peregrine Capital Limited J&A Securities (Hong Kong) Limited Shanghai International Capital (H.K.) Limited Wheelock NatWest Securities Limited Seapower Securities Limited Amsteel Securities (H.K.) Limited China Southern Corporate Finance Ltd. -
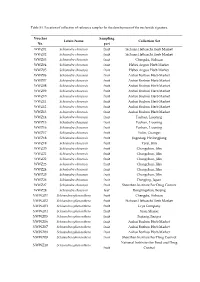
Table S1. Location of Collection of Reference Samples for the Development of the Nucleotide Signature
Table S1. Location of collection of reference samples for the development of the nucleotide signature. Voucher Sampling Latain Name Collection Set No. part WWZ01 Schisandra chinensis fruit Sichuan Hehuachi Herb Market WWZ02 Schisandra chinensis fruit Sichuan Hehuachi Herb Market WWZ03 Schisandra chinensis fruit Chengdu, Sichuan WWZ04 Schisandra chinensis fruit Hebei Anguo Herb Market WWZ05 Schisandra chinensis fruit Hebei Anguo Herb Market WWZ06 Schisandra chinensis fruit Anhui Bozhou Herb Market WWZ07 Schisandra chinensis fruit Anhui Bozhou Herb Market WWZ08 Schisandra chinensis fruit Anhui Bozhou Herb Market WWZ09 Schisandra chinensis fruit Anhui Bozhou Herb Market WWZ10 Schisandra chinensis fruit Anhui Bozhou Herb Market WWZ11 Schisandra chinensis fruit Anhui Bozhou Herb Market WWZ12 Schisandra chinensis fruit Anhui Bozhou Herb Market WWZ13 Schisandra chinensis fruit Anhui Bozhou Herb Market WWZ14 Schisandra chinensis fruit Fushun, Liaoning WWZ15 Schisandra chinensis fruit Fushun, Liaoning WWZ16 Schisandra chinensis fruit Fushun, Liaoning WWZ17 Schisandra chinensis fruit Yulin, Guangxi WWZ18 Schisandra chinensis fruit Jiagedaqi, Heilongjiang WWZ19 Schisandra chinensis fruit Yanji, Jilin WWZ20 Schisandra chinensis fruit Changchun, Jilin WWZ21 Schisandra chinensis fruit Changchun, Jilin WWZ22 Schisandra chinensis fruit Changchun, Jilin WWZ23 Schisandra chinensis fruit Changchun, Jilin WWZ24 Schisandra chinensis fruit Changchun, Jilin WWZ25 Schisandra chinensis fruit Changchun, Jilin WWZ26 Schisandra chinensis fruit Dongjing, Japan WWZ27 -

Clean Urban/Rural Heating in China: the Role of Renewable Energy
Clean Urban/Rural Heating in China: the Role of Renewable Energy Xudong Yang, Ph.D. Chang-Jiang Professor & Vice Dean School of Architecture Tsinghua University, China Email: [email protected] September 28, 2020 Outline Background Heating Technologies in urban Heating Technologies in rural Summary and future perspective Shares of building energy use in China 2018 Total Building Energy:900 million tce+ 90 million tce biomass 2018 Total Building Area:58.1 billion m2 (urban 34.8 bm2 + rural 23.3 bm2 ) Large-scale commercial building 0.4 billion m2, 3% Normal commercial building Rural building 4.9 billion m2, 18% 24.0 billion m2, 38% Needs clean & efficient Space heating in North China (urban) 6.4 billion m2, 25% Needs clean and efficient Heating of residential building Residential building in the Yangtze River region (heating not included) 4.0 billion m2, 1% 9.6 billion m2, 15% Different housing styles in urban/rural Typical house (Northern rural) Typical housing in urban areas Typical house (Southern rural) 4 Urban district heating network Heating terminal Secondary network Heat Station Power plant/heating boiler Primary Network Second source Pump The role of surplus heat from power plant The number Power plant excess of prefecture- heat (MW) level cities Power plant excess heat in northern China(MW) Daxinganling 0~500 24 500~2000 37 Heihe Hulunbier 2000~5000 55 Yichun Hegang Tacheng Aletai Qiqihaer Jiamusi Shuangyashan Boertala Suihua Karamay Qitaihe Jixi 5000~10000 31 Xingan Daqing Yili Haerbin Changji Baicheng Songyuan Mudanjiang