Genetic Diversity Among Isolates of Phytophthora Sojae in Anhui Province of China Based on ISSR- PCR Markers
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Research Article Stability and Complexity Analysis of Temperature Index Model Considering Stochastic Perturbation
Hindawi Advances in Mathematical Physics Volume 2018, Article ID 2789412, 18 pages https://doi.org/10.1155/2018/2789412 Research Article Stability and Complexity Analysis of Temperature Index Model Considering Stochastic Perturbation Jing Wang Faculty of Science, Bengbu University, Bengbu 233030, China Correspondence should be addressed to Jing Wang; [email protected] Received 20 October 2017; Accepted 12 December 2017; Published 1 January 2018 Academic Editor: Giampaolo Cristadoro Copyright © 2018 Jing Wang. Tis is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. A temperature index model with delay and stochastic perturbation is constructed in this paper. It explores the infuence of parameters and stochastic factors on the stability and complexity of the model. Based on historical temperature data of four cities of Anhui Province in China, the temperature periodic variation trends of approximately sinusoidal curves of four cities are given, respectively. In addition, we analyze the existence conditions of the local stability of the temperature index model without stochastic term and estimate its parameters by using the same historical data of the four cities, respectively. Te numerical simulation results of the four cities are basically consistent with the descriptions of their historical temperature data, which proves that the temperature index model constructed has good ftting degree. It also shows that unreasonable delay parameter can make the model lose stability and improve the complexity. Stochastic factors do not usually change the trend in temperature, but they can cause high frequency fuctuations in the process of temperature evolution. -

Procured Contracts February/13
Implementation Schedule of Remaining Contracts Not Procured Yet Public Disclosure Authorized 1USD=6.3 CNY (based on the exchange rate as of Nov, 2012) February/13 Estimated Base Costs Expected Comp- Sub- Procure- Prequa- Review by Expected Contract Contract Construction No. onent Component component Sub-component Contract Name ment lifica- WB (Prior Bid Opening Remark Package No. Type Duration No. No. Mothod tion /Post) Date CNY10,000 USD10,000 (month) Wuxiaojie Storm Drainage Pump Civil 1 HSFS/S5/C1 7200.00 1142.86 NCB NO Prior Jan/13 17 in the bidding Huaishang Station & Associated Works Suburban District Flood Public Disclosure Authorized Works Environment Management & 3 Infrastructure 3 Storm Drainage Improvement and Infrastructure Reconstruction & Improvement Expansion of HSFS/S5/C6-N- Wangxiaogou Pump Civil 2 3478.00 552.06 NCB NO Post Apr/13 16 B Stations and Works Wangxiaogou Intake Ditch Public Disclosure Authorized Lilou Road (Donghai New Addition Civil 4 LZUI/S10/C1 Avenue - Huangshan 10572.00 1678.10 NCB NO Prior Apr/13 12 Project of Mid- Works Urban Urban(south of Avenue) Term Adjustment Environment Huai River) 2 3 Infrastructure Infrastructure Fengandong Road Improvement Improvement New Addition (High Speed Rail Civil 5 LZUI/S10/C2 10351.00 1643.02 NCB NO Prior Apr/13 12 Project of Mid- Culvert - Mid. Ring Works Term Adjustment Road) Civil Works Contracts Total 31601.00 5016.03 Public Disclosure Authorized Preparation of Rules and Operational Procedures for Mohekou Industrial Bid evaluation 6 C Zone (MIZ), and - 20 TA QCBS - Prior Aug/12 12 is in process Bidding Document to Select a Professional Technical Operator for MIZ Services, 4 Training and Study Tour Institutional & Financial Bid evaluation 7 E - 32 TA QBS - Prior Aug/12 12 Strengthening of is in process Utility Companies Strategy and Selection of Bengbu 8 F - 21 TA CQS - Prior Mar/13 7 Water Sector Development TA and Training Total - 73 Procured Contracts February/13 Review Estimated Cost Cost of Signed Contract Procure- Contract by WB Bid Opening Contract No. -

Analysis of Historic Rainfall Characteristics for Robust Wheat Cropping in North Anhui
Analysis of historic rainfall characteristics for robust wheat cropping in North Anhui Hui Su, Yibo Wu, Yulei Zhu, Youhong Song Anhui Agricultural University, School of Agronomy, Hefei, 233036. Corresponding email: [email protected] Abstract Northern part of Anhui is one of major wheat producing areas in China. The total amount of rainfall is sufficient for wheat season; however, it is unevenly distributed at the different growth stages, resulting in risk of yield losses. In order to optimise the cultivation in North Anhui, it is essential to characterise the rainfall pattern for wheat growth particularly in the critical period (i.e. the months of sowing and harvesting). By analysing the rainfall data from 1955 to 2017, this study characterised the rainfall pattern from six sites representing different regions of North Anhui. The frequency of continuous rainfall days during sowing and harvesting periods were quantified based on 63 years rainfall distribution. The characterisation of rainfall in six representative sites in North Anhui were able to be used to guide wheat sowing and harvesting, which could help farmers to make decisions and avoid likelihood of cropping risks. Key Words Winter wheat, rainfall pattern, sustainable cropping, China Introduction Huang-Huai-Hai plain is a major wheat production area in China (Yang, 2018). The northern part of Anhui province belongs to south Huang-Huai-Hai plain. The wheat sowing time normally varies 1 week before or after October 15 and harvesting is tightly closing to May 31 in the following year. The climate characteristics particularly rainfall distribution over the season are complicated in North Anhui as it is in the transitional zone between the North and South China. -

World Bank Document
CONFORMED COPY LOAN NUMBER 4597 CHA Public Disclosure Authorized Loan Agreement (Huai River Pollution Control Project) between PEOPLE’S REPUBLIC OF CHINA and Public Disclosure Authorized INTERNATIONAL BANK FOR RECONSTRUCTION AND DEVELOPMENT Dated September 24, 2001 LOAN NUMBER 4597 CHA Public Disclosure Authorized LOAN AGREEMENT AGREEMENT, dated September 24, 2001, between PEOPLE'S REPUBLIC OF CHINA (the Borrower) and INTERNATIONAL BANK FOR RECONSTRUCTION AND DEVELOPMENT (the Bank). WHEREAS (A) the Borrower, having satisfied itself as to the feasibility and priority of the project described in Schedule 2 to this Agreement (the Project), has requested the Bank to assist in the financing of the Project; (B) the Project will be carried out by the Borrower's Provinces of Anhui and Shandong (collectively, the Project Provinces and individually, a Project Province) with the Borrower’s assistance and, as part of such assistance, the Borrower will make the proceeds of the loan provided for in Article II of this Agreement (the Loan) available to each of the Project Provinces, as set forth in this Agreement; and WHEREAS the Bank has agreed, on the basis, inter alia, of the foregoing, to extend the Loan to the Borrower upon the terms and conditions set forth in this Agreement and in the Project Agreement of even date herewith between the Bank and the Public Disclosure Authorized Project Provinces (the Project Agreement); NOW THEREFORE the parties hereto hereby agree as follows: ARTICLE I General Conditions; Definitions Section 1.01. The "General Conditions Applicable to Loan and Guarantee Agreements for Single Currency Loans" of the Bank, dated May 30, 1995, as amended through October 6, 1999 (the General Conditions), constitute an integral part of this Agreement. -

Study on the Spatial-Temporal Pattern Evolution and Countermeasures of Regional Coordinated Development in Anhui Province, China
Current Urban Studies, 2020, 8, 115-128 https://www.scirp.org/journal/cus ISSN Online: 2328-4919 ISSN Print: 2328-4900 Study on the Spatial-Temporal Pattern Evolution and Countermeasures of Regional Coordinated Development in Anhui Province, China Yizhen Zhang1,2*, Weidong Cao1,2, Kun Zhang3 1School of Geography and Tourism, Anhui Normal University, Wuhu, China 2Urban and Regional Planning Research Center of Anhui Normal University, Wuhu, China 3College of Tourism, Huaqiao University, Quanzhou, China How to cite this paper: Zhang, Y. Z., Cao, Abstract W. D., & Zhang, K. (2020). Study on the Spatial-Temporal Pattern Evolution and Regional coordinated development was an important measure to resolve new Countermeasures of Regional Coordinated contradictions in the new era, and it has gradually become a hotspot in geog- Development in Anhui Province, China. raphy research. At three time points of 2010, 2013, and 2017, by constructing Current Urban Studies, 8, 115-128. an evaluation index system for the city comprehensive competitiveness, the https://doi.org/10.4236/cus.2020.81005 entropy method and the coupling coordination model were used to study the Received: March 2, 2020 coordinated development pattern of cities in Anhui Province. Moreover, we Accepted: March 27, 2020 tried to raise the issue of regional coordinated development in Anhui Prov- Published: March 30, 2020 ince and corresponding countermeasures. The results showed that the com- Copyright © 2020 by author(s) and prehensive competitiveness of cities in Anhui Province has strong spatio- Scientific Research Publishing Inc. temporal heterogeneity. The spatial development pattern with Hefei as the This work is licensed under the Creative core was more obvious. -

Quantitative Analysis of Burden of Infectious Diarrhea Associated with Floods in Northwest of Anhui Province, China: a Mixed Method Evaluation
Quantitative Analysis of Burden of Infectious Diarrhea Associated with Floods in Northwest of Anhui Province, China: A Mixed Method Evaluation Guoyong Ding1, Ying Zhang2, Lu Gao1, Wei Ma1, Xiujun Li1, Jing Liu1, Qiyong Liu3, Baofa Jiang1* 1 Department of Epidemiology and Health Statistics, School of Public Health, Shandong University, Jinan City, Shandong Province, P.R. China, 2 School of Public Health, China Studies Centre, The University of Sydney, New South Wales, Australia, 3 National Institute for Communicable Disease Control and Prevention, China CDC, Beijing City, P.R. China Abstract Background: Persistent and heavy rainfall in the upper and middle Huaihe River of China brought about severe floods during the end of June and July 2007. However, there has been no assessment on the association between the floods and infectious diarrhea. This study aimed to quantify the impact of the floods in 2007 on the burden of disease due to infectious diarrhea in northwest of Anhui Province. Methods: A time-stratified case-crossover analysis was firstly conducted to examine the relationship between daily cases of infectious diarrhea and the 2007 floods in Fuyang and Bozhou of Anhui Province. Odds ratios (ORs) of the flood risk were quantified by conditional logistic regression. The years lived with disability (YLDs) of infectious diarrhea attributable to floods were then estimated based on the WHO framework of the calculating potential impact fraction in the Burden of Disease study. Results: A total of 197 infectious diarrheas were notified during the exposure and control periods in the two study areas. The strongest effect was shown with a 2-day lag in Fuyang and a 5-day lag in Bozhou. -

Quantitative Assessment and Diagnosis for Regional Agricultural Drought Resilience Based on Set Pair Analysis and Connection Entropy
Article Quantitative Assessment and Diagnosis for Regional Agricultural Drought Resilience Based on Set Pair Analysis and Connection Entropy Menglu Chen 1, Shaowei Ning 1, Yi Cui 2,*, Juliang Jin 1, Yuliang Zhou 1 and Chengguo Wu 1 1 School of Civil Engineering, Hefei University of Technology, Hefei 230009, China; [email protected] (M.C.); [email protected] (S.N.); [email protected] (J.J.); [email protected] (Y.Z.); [email protected] (C.W.) 2 State Key Laboratory of Hydraulic Engineering Simulation and Safety, Tianjin University, Tianjin 300072, China * Correspondence: [email protected]; Tel.: +86-13102218320 Received: 27 February 2019; Accepted: 02 April 2019; Published: 5 April 2019 Abstract:Assessment and diagnosis of regional agricultural drought resilience (RADR) is an important groundwork to identify the shortcomings of regional agriculture to resist drought disasters accurately. In order to quantitatively assess the capacity of regional agriculture system to reduce losses from drought disasters under complex conditions and to identify vulnerability indexes, an assessment and diagnosis model for RADR was established. Firstly, this model used the improved fuzzy analytic hierarchy process to determine the index weights, then proposed an assessment method based on connection number and an improved connection entropy. Furthermore, the set pair potential based on subtraction was used to diagnose the vulnerability indexes. In addition, a practical application had been carried out in the region of the Huaibei Plain in Anhui Province. The evaluation results showed that the RADR in this area from 2005 to 2014 as a whole was in a relatively weak situation. However, the average grade values had decreased from 3.144 to 2.790 during these 10 years and the RADR had an enhanced tendency. -

Online Appendix (474.67
How do Tax Incentives Aect Investment and Productivity? Firm-Level Evidence from China ONLINE APPENDIX Yongzheng Liu School of Finance Renmin University of China E-mail: [email protected] Jie Mao School of International Trade and Economics University of International Business and Economics E-mail: [email protected] 1 Appendix A: Supplementary Figures and Tables Figure A1: The Distribution of Estimates for the False VAT Reform Variable Panel A. ln(Investment) Panel B. ln(TFP, OP method) 15 50 40 10 30 20 5 Probabilitydensity Probability density 10 0 0 -0.10 0.00 0.10 0.384 -0.02 0.00 0.02 0.089 The simulated VAT reform estimate The simulated VAT reform estimate reference normal, mean .0016 sd .03144 reference normal, mean .00021 sd .00833 Notes: The gure plots the density of the estimated coecients of the false VAT reform variable from the 500 simulation tests using the specication in Column (3) of Table 2. The vertical red lines present the treatment eect estimates reported in Column (3) of Table 2. Source: Authors' calculations. 2 Table A1: Evolution of the VAT Reform in China Stage of the Reform Industries Covered (Industry Classication Regions Covered (Starting Codes) Time) Machine and equipment manufacturing (35, 36, 39, 40, 41, 42); Petroleum, chemical, and pharmaceutical manufacturing (25, 26, 27, 28, 29, 30); Ferrous and non-ferrous metallurgy (32, 33); The three North-eastern provinces: Liaoning (including 1 (July 2004) Agricultural product processing (13, 14, 15, 17, 18, 19, 20, 21, Dalian city), Jilin and Heilongjiang. 22); Shipbuilding (375); Automobile manufacturing (371, 372, 376, 379); Selected military and hi-tech products (a list of 249 rms, 62 of which are in our sample). -

Status of Aphanomyces Root Rot in Wisconsin
STATUS OF APHANOMYCES ROOT ROT IN WISCONSIN C.R. Grau1 Introduction Alfalfa is the primary forage crop in Wisconsin and is a key element in the state’s dairy industry. The yield of new varieties is greater than that of Vernal and other older varieties due to genetic gains made by breeders over the years. Much of the yield advantage of new varieties may be attributed to efforts to breed for resistance to a wide variety of major pathogens of alfalfa such as Verticillium, Phytophthora, and Aphanomyces. Although the yield gap has widened between new varieties and Vernal, yield of all varieties has declined steadily in Wisconsin during the past 30 years (Wiersma et al., 1997). There are several possible explanations for this situation including changing climate and the difficulties inherent in dealing with such a genetically diverse crop such as alfalfa. From a pathologist’s perspective, new disease-causing organisms or new strains of previously described pathogens may also play a role in limiting yield gains for Wisconsin alfalfa growers. At the University of Wisconsin-Madison, we are conducting research on alfalfa diseases, particularly those caused by organisms that have not been studied previously with respect to their presence and influence on the alfalfa crop. Aphanomyces root rot is one relatively new disease of alfalfa that has been studied in this regard. Overview of Aphanomyces Root Rot Although the cause, the fungus Aphanomyces euteiches, was reported to infect alfalfa in the 1930’s, this organism was considered a minor pathogen of alfalfa. In contrast, Phytophthora root rot, caused by Phytophthora medicaginis, was considered the only significant cause of root disease of alfalfa in wet soil situations. -

PRC: Anhui Integrated Transport Sector Improvement Project–Xuming Expressway
Updated Resettlement Plan January 2011 PRC: Anhui Integrated Transport Sector Improvement Project–Xuming Expressway Prepared by Anhui Provincial Communications Investment Group Company for the Asian Development Bank. 2 CURRENCY EQUIVALENTS Currency unit – Yuan (CNY) $1.00 = CNY6.80 ABBREVIATIONS ACTVC - Anhui Communications Vocational & Technical College ADB - Asian Development Bank AHAB - Anhui Highway Administration Bureau APCD - Anhui Provincial Communications Department Anhui Provincial Communications Investment Group ACIG - Company APs Affected Persons AVs Affected Villages APG - Anhui Provincial Government M&E Monitoring and Evaluation PMO - Project Management Office RP - Resettlement Plan PRC - People’s Republic of China NOTE (i) In this report, "$" refers to US dollars unless otherwise stated. This updated resettlement plan is a document of the borrower. The views expressed herein do not necessarily represent those of ADB's Board of Directors, Management, or staff, and may be preliminary in nature. Your attention is directed to the “terms of use” section of this website. In preparing any country program or strategy, financing any project, or by making any designation of or reference to a particular territory or geographic area in this document, the Asian Development Bank does not intend to make any judgments as to the legal or other status of any territory or area. ADB Financed Anhui Integrated Transport Sector Improvement Project Resettlement Plan for Anhui Xuzhou-Mingguang Expressway Project (updated) Anhui Provincial Communications Investment Group Company January, 2011 Note on the updated RP On November 11, 2009, the Anhui Provincial Development and Reform Commission gave a reply on the detailed design of the Anhui section of the Xuzhou-Mingguang Expressway with Document APDRC [2009] No.1199. -
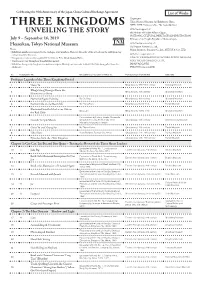
Three Kingdoms Unveiling the Story: List of Works
Celebrating the 40th Anniversary of the Japan-China Cultural Exchange Agreement List of Works Organizers: Tokyo National Museum, Art Exhibitions China, NHK, NHK Promotions Inc., The Asahi Shimbun With the Support of: the Ministry of Foreign Affairs of Japan, NATIONAL CULTURAL HERITAGE ADMINISTRATION, July 9 – September 16, 2019 Embassy of the People’s Republic of China in Japan With the Sponsorship of: Heiseikan, Tokyo National Museum Dai Nippon Printing Co., Ltd., Notes Mitsui Sumitomo Insurance Co.,Ltd., MITSUI & CO., LTD. ・Exhibition numbers correspond to the catalogue entry numbers. However, the order of the artworks in the exhibition may not necessarily be the same. With the cooperation of: ・Designation is indicated by a symbol ☆ for Chinese First Grade Cultural Relic. IIDA CITY KAWAMOTO KIHACHIRO PUPPET MUSEUM, ・Works are on view throughout the exhibition period. KOEI TECMO GAMES CO., LTD., ・ Exhibition lineup may change as circumstances require. Missing numbers refer to works that have been pulled from the JAPAN AIRLINES, exhibition. HIKARI Production LTD. No. Designation Title Excavation year / Location or Artist, etc. Period and date of production Ownership Prologue: Legends of the Three Kingdoms Period 1 Guan Yu Ming dynasty, 15th–16th century Xinxiang Museum Zhuge Liang Emerges From the 2 Ming dynasty, 15th century Shanghai Museum Mountains to Serve 3 Narrative Figure Painting By Qiu Ying Ming dynasty, 16th century Shanghai Museum 4 Former Ode on the Red Cliffs By Zhang Ruitu Ming dynasty, dated 1626 Tianjin Museum Illustrated -

Research on Logistics Efficiency of Anhui Province
2019 4th International Workshop on Materials Engineering and Computer Sciences (IWMECS 2019) Research on Logistics Efficiency of Anhui Province Han Yanhui School of Traffic and Transportation, Beijing Jiaotong University, P. R. China [email protected] Keywords: Logistics efficiency; DEA model; Anhui province Abstract: China's logistics industry has developed rapidly in recent years, and logistics efficiency is the main indicator that reflects the level of logistics development. This article first illustrated the situation of logistics development in Anhui Province and established an index system for evaluation of logistics efficiency, including four input indicators, the number of employees in the logistics industry, the mileage of graded roads, investment in fixed assets in the logistics industry and education expenditure, as well as two output indicators, the freight turnover volume and the added value of the logistics industry; then use the CCR-DEA and BCC-DEA models to evaluate the logistics efficiency of 16 cities of Anhui province. The results show that there is a phenomenon of unbalanced logistics development. Among them, Suzhou, Chizhou, and Anqing have low logistics efficiency, which is the bottleneck that limits the improvement of logistics efficiency in Anhui Province. 1. Introduction With the growth of China's economic level and the continuous transformation of economic growth pattern, the development level of logistics industry has become an important index to measure the comprehensive development level of a region. Anhui province is located in the middle and lower reaches of the Yangtze river and the Huaihe river. Together with Jiangsu, Shanghai and Zhejiang, forms the Yangtze river delta urban agglomeration.