Molecular Evolutionary Relationships in the Avian Genus Anthus (Pipits
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
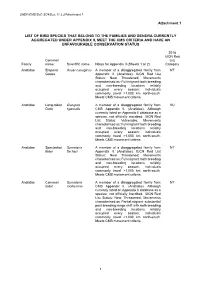
Attachment 1 LIST of BIRD SPECIES THAT BELONG to the FAMILIES
UNEP/CMS/ScC-SC4/Doc.11.3.2/Attachment 1 Attachment 1 LIST OF BIRD SPECIES THAT BELONG TO THE FAMILIES AND GENERA CURRENTLY AGGREGATED UNDER APPENDIX II, MEET THE CMS CRITERIA AND HAVE AN UNFAVOURABLE CONSERVATION STATUS 2018 IUCN Red Common List Family name Scientific name Notes for Appendix II (Sheets 1 or 2) Category Anatidae Emperor Anser canagicus A member of a disaggregated family from NT Goose Appendix II. (Anatidae). IUCN Red List Status: Near Threatened; Movements characterised as: Full migrant: both breeding and non-breeding locations reliably occupied every season; individuals commonly travel >1,000 km north-south. Meets CMS movement criteria. Anatidae Long-tailed Clangula A member of a disaggregated family from VU Duck hyemalis CMS Appendix II. (Anatidae). Although currently listed on Appendix II database as a species, not officially inscribed. IUCN Red List Status: Vulnerable; Movements characterised as: Full migrant: both breeding and non-breeding locations reliably occupied every season; individuals commonly travel >1,000 km north-south. Meets CMS movement criteria. Anatidae Spectacled Somateria A member of a disaggregated family from NT Eider fischeri Appendix II. (Anatidae). IUCN Red List Status: Near Threatened; Movements characterised as: Full migrant: both breeding and non-breeding locations reliably occupied every season; individuals commonly travel >1,000 km north-south. Meets CMS movement criteria. Anatidae Common Somateria A member of a disaggregated family from NT Eider mollissima CMS Appendix II. (Anatidae). Although currently listed on Appendix II database as a species, not officially inscribed. IUCN Red List Status: Near Threatened; Movements characterised as: Partial migrant: substantial post-breeding range shift with both breeding and non-breeding locations reliably occupied every season; individuals commonly travel >1,000 km north-south. -

Predicting the Distribution of the Vulnerable Yellow-Breasted Pipit (Anthus Chloris) Using Species Distribution Modelling
1 Predicting the distribution of the Vulnerable Yellow-breasted Pipit (Anthus chloris) using Species Distribution Modelling Darren W. Pietersen1*, Ian T. Little2, Raymond Jansen3 and Andrew E. McKechnie1 1DST-NRF Centre of Excellence at the Percy FitzPatrick Institute, Department of Zoology and Entomology, University of Pretoria, Private Bag X20, Hatfield, 0028, South Africa 2The Endangered Wildlife Trust, Private Bag X11, Modderfontein, 1645, Johannesburg, South Africa 3Department of Environmental, Water and Earth Sciences, Tshwane University of Technology, Private Bag X680, Pretoria, 0001, South Africa *Address for correspondence: Department of Zoology and Entomology University of Pretoria Private Bag X20 Hatfield 0028 [email protected] +27 82 937 6052 2 Abstract The Yellow-breasted Pipit (Anthus chloris) is endemic to the eastern escarpment of South Africa, marginally entering eastern Lesotho. This species is classified as globally Vulnerable due to a perceived decreasing population size and loss of habitat. We employed Species Distribution Modelling to investigate the predicted range of this species to determine whether additional purportedly suitable habitat exists where this species may be found, and to assess the degree to which habitat loss has affected this species. We used a database of 250 independently obtained and verified sightings to predict the summer breeding distribution of this species and compare our verified sightings and predicted range to the sightings currently in the Second Southern African Bird Atlas Project (SABAP2) database and the latest regional conservation assessment. Our models closely approximate the current distribution of the Yellow-breasted Pipit, and suggest that most of the purportedly suitable habitat is occupied, at least at the macro-scale. -

Bird Checklists of the World Country Or Region: Myanmar
Avibase Page 1of 30 Col Location Date Start time Duration Distance Avibase - Bird Checklists of the World 1 Country or region: Myanmar 2 Number of species: 1088 3 Number of endemics: 5 4 Number of breeding endemics: 0 5 Number of introduced species: 1 6 7 8 9 10 Recommended citation: Lepage, D. 2021. Checklist of the birds of Myanmar. Avibase, the world bird database. Retrieved from .https://avibase.bsc-eoc.org/checklist.jsp?lang=EN®ion=mm [23/09/2021]. Make your observations count! Submit your data to ebird. -

India: Kaziranga National Park Extension
INDIA: KAZIRANGA NATIONAL PARK EXTENSION FEBRUARY 22–27, 2019 The true star of this extension was the Indian One-horned Rhinoceros (Photo M. Valkenburg) LEADER: MACHIEL VALKENBURG LIST COMPILED BY: MACHIEL VALKENBURG VICTOR EMANUEL NATURE TOURS, INC. 2525 WALLINGWOOD DRIVE, SUITE 1003 AUSTIN, TEXAS 78746 WWW.VENTBIRD.COM INDIA: KAZIRANGA NATIONAL PARK EXTENSION February 22–27, 2019 By Machiel Valkenburg This wonderful Kaziranga extension was part of our amazing Maharajas’ Express train trip, starting in Mumbai and finishing in Delhi. We flew from Delhi to Guwahati, located in the far northeast of India. A long drive later through the hectic traffic of this enjoyable country, we arrived at our lodge in the evening. (Photo by tour participant Robert Warren) We enjoyed three full days of the wildlife and avifauna spectacles of the famous Kaziranga National Park. This park is one of the last easily accessible places to find the endangered Indian One-horned Rhinoceros together with a healthy population of Asian Elephant and Asiatic Wild Buffalo. We saw plenty individuals of all species; the rhino especially made an impression on all of us. It is such an impressive piece of evolution, a serious armored “tank”! On two mornings we loved the elephant rides provided by the park; on the back of these attractive animals we came very close to the rhinos. The fertile flood plains of the park consist of alluvial silts, exposed sandbars, and riverine flood-formed lakes called Beels. This open habitat is not only good for mammals but definitely a true gem for some great birds. Interesting but common birds included Bar-headed Goose, Red Junglefowl, Woolly-necked Stork, and Lesser Adjutant, while the endangered Greater Adjutant and Black-necked Stork were good hits in the stork section. -

Status of Red-Throated Pipit Anthus Cervinus in Bangladesh
104 SHORT NOTES Forktail 27 (2011) Acknowledgements Lim, K. S. (2009) The avifauna of Singapore . Singapore: Nature Society I thank Alfred Chia, Lee Tiah Khee, Lim Kim Chuah and Lim Kim Seng for (Singapore). generously sharing their observations. Thanks also go to Kelvin Lim and Tan Ng, H. H. & Tan, H. H. (2010) An annotated checklist of the non-native freshwater Heok Hui at the Raffles Museum of Biodiversity Research, and Albert Low in fish species in the reservoirs of Singapore. Cosmos 6(1): 95–116. helping to identify fish species. I am grateful to Ruth Tingay and Benjamin Remahl, U. (2008) Grey-headed fish-eagle at “Little Guilin”. Singapore Avifauna Lee for providing useful inputs during the preparation of this manuscript. 22(7): 9–10. Lastly, I thank Todd E. Katzner and Clive Briffett for their comments which Sergio, F., Caro, T., Brown, D., Clucas, B., Hunter, J., Ketchum, J., McHugh, K. & improved the manuscript greatly. Hirauldo, F. (2008) Top predators as conservation tools: ecological rationale, assumptions and efficacy. Ann. Rev. Ecol. Evol. & Systematics 39: References 1–19. Baker, N. & Lim, K. (2008) Wild animals of Singapore . Singapore: Draco Tingay, R. E., Nicoll, M. A. C. & Sun Visal (2006) Status and distribution of the Publishing Pte Ltd and Nature Society (Singapore). grey-headed fish-eagle ( Ichthyophaga ichthyaetus ) in the Prek Toal core Birdlife International (2011) Species factsheet: Ichthyophaga ichthyaetus . area of Tonle Sap lake, Cambodia. J. Raptor Res. 40: 277–283. Downloaded from http://www.birdlife.org on 20/01/2011. Tingay, R. E., Nicoll, M. A. C., Whitfield, D. -

Red List of Bangladesh 2015
Red List of Bangladesh Volume 1: Summary Chief National Technical Expert Mohammad Ali Reza Khan Technical Coordinator Mohammad Shahad Mahabub Chowdhury IUCN, International Union for Conservation of Nature Bangladesh Country Office 2015 i The designation of geographical entitles in this book and the presentation of the material, do not imply the expression of any opinion whatsoever on the part of IUCN, International Union for Conservation of Nature concerning the legal status of any country, territory, administration, or concerning the delimitation of its frontiers or boundaries. The biodiversity database and views expressed in this publication are not necessarily reflect those of IUCN, Bangladesh Forest Department and The World Bank. This publication has been made possible because of the funding received from The World Bank through Bangladesh Forest Department to implement the subproject entitled ‘Updating Species Red List of Bangladesh’ under the ‘Strengthening Regional Cooperation for Wildlife Protection (SRCWP)’ Project. Published by: IUCN Bangladesh Country Office Copyright: © 2015 Bangladesh Forest Department and IUCN, International Union for Conservation of Nature and Natural Resources Reproduction of this publication for educational or other non-commercial purposes is authorized without prior written permission from the copyright holders, provided the source is fully acknowledged. Reproduction of this publication for resale or other commercial purposes is prohibited without prior written permission of the copyright holders. Citation: Of this volume IUCN Bangladesh. 2015. Red List of Bangladesh Volume 1: Summary. IUCN, International Union for Conservation of Nature, Bangladesh Country Office, Dhaka, Bangladesh, pp. xvi+122. ISBN: 978-984-34-0733-7 Publication Assistant: Sheikh Asaduzzaman Design and Printed by: Progressive Printers Pvt. -

Record of Slaty-Breasted Rail Rallus Striatus Breeding in Dehradun, Indiap Kumar, RS Kumar
Indian Birds Vol. 5 No. 1 ISSN 0973-1407 Editor Emeritus Zafar Futehally Editor Contents Aasheesh Pittie Email: [email protected] Management of Greater Short-toed Larks Associate Editor V. Santharam Calandrella brachydactyla in Indian aerodromes S. S. Mahesh 2 Editorial Board Maan Barua Observations on the globally threatened Pied Tit Anwaruddin Choudhury Parus nuchalis at Jessore Sanctuary, Gujarat Bill Harvey Pranav Trivedi 7 Farah Ishtiaq Rajah Jayapal The raptors and the agamid Madhusudan Katti Ashwin Baindur 11 R. Suresh Kumar Taej Mundkur Two new birds for Gujarat Rishad Naoroji Uffe Gjøl Sørensen & Jugal K. Tiwari 14 Suhel Quader Harkirat Singh Sangha Short News C. Sashikumar A substitute name for Parus nipalensis Hodgson, 1837 S. Subramanya Edward C. Dickinson 16 K. Gopi Sundar Record of the Great White Pelican Pelecanus onocrotalus from Uppalapadu, Contributing Editors Andhra Pradesh, India Praveen J. Nanjan Sheeba & Lalitha Vijayan 17 Ragupathy Kannan Lavkumar Khachar Breeding of Glossy Ibis Plegadis falcinellus at Contributing Photographer Vedanthangal Waterbird Sanctuary, southern India Clement Francis C. Venkatraman 18 Layout & Design The Black Baza Aviceda leuphotes in Chennai: a review of sight records K. Jayaram V. Santharam 19 Office Record of Slaty-breasted Rail Rallus striatus breeding in Dehradun, India P. Rambabu Pankaj Kumar & R. Suresh Kumar 21 Common Moorhen Gallinula chloropus in the diet of the African Catfish NEW ORNIS FOUNDATION Clarias gariepinus in Keoladeo Ghana National Park, India Registration No. 314/2004 K. R. Anoop, K. S. Gopi Sundar, Bholu Abrar Khan & Sohan Lal 22 URL: www.indianbirds.in The first breeding record of the Asian Brown Flycatcher Muscicapa dauurica poonensis Trustees in Rajasthan, India Zafar Futehally Harkirat Singh Sangha, Gobind Sagar Bhardwaj & Devender Mistry 24 Aasheesh Pittie V. -

The Birds of Okhla Barrage Bird Sanctuary, Delhi, India
FORKTAIL 19 (2003): 39-50 The birds of Okhla barrage bird sanctuary, Delhi, India ABDUL JAMIL URFI Okhla barrage bird sanctuary, on the river Yamuna in south Delhi, is an important site for breeding and wintering waterbirds, with 14,000–20,000 waterbirds recorded in winter. The dominant feature of the site is a large lake formed after the creation of a barrage on the river in 1986. Historical records, records resulting from fieldwork since 1989, and other recent records have been combined to produce a list of 302 species for the site and its immediate area. Species recorded since 1992 include three Vulnerable species (Baer’s Pochard Aythya baeri, Indian Skimmer Rynchops albicollis and Bristled Grassbird Chaetornis striatus) and six Near Threatened species (Ferruginous Pochard Aythya nyroca, Black-bellied Tern Sterna acuticauda, Darter Anhinga melanogaster, Black-headed Ibis Threskiornis melanocephalus, Painted Stork Mycteria leucocephala and Black-necked Stork Ephippiorhynchus asiaticus). The sanctuary is being encroached upon, and isolated by, surrounding development. INTRODUCTION The Okhla barrage bird sanctuary in Delhi is a haven for waterbirds (Singh 1983, Urfi 1993a, b, 1995). In 1990, an area of c.3.5 km2 on the river Yamuna in Delhi was notified as a bird sanctuary by the Uttar Pradesh government under the Wildlife Protection Act of India. The site is located at the point where the river leaves the territory of Delhi and enters the neighbouring state of Uttar Pradesh (Fig. 1). The most prominent feature of the sanctuary is the large lake created by damming the river, which lies sandwiched between Okhla village towards the west and Gautambudh Nagar towards the east. -

Multi-Locus Phylogeny of African Pipits and Longclaws (Aves: Motacillidae) Highlights Taxonomic Inconsistencies
Running head: African pipit and longclaw taxonomy Multi-locus phylogeny of African pipits and longclaws (Aves: Motacillidae) highlights taxonomic inconsistencies DARREN W. PIETERSEN,1* ANDREW E. MCKECHNIE,1,2 RAYMOND JANSEN,3 IAN T. LITTLE4 AND ARMANDA D.S. BASTOS5 1DST-NRF Centre of Excellence at the Percy FitzPatrick Institute, Department of Zoology and Entomology, University of Pretoria, Hatfield, South Africa 2South African Research Chair in Conservation Physiology, National Zoological Garden, South African National Biodiversity Institute, P.O. Box 754, Pretoria 0001, South Africa 3Department of Environmental, Water and Earth Sciences, Tshwane University of Technology, Pretoria, South Africa 4Endangered Wildlife Trust, Johannesburg, South Africa 5Department of Zoology and Entomology, University of Pretoria, Hatfield, South Africa *Corresponding author. Email: [email protected] 1 Abstract The globally distributed avian family Motacillidae consists of 5–7 genera (Anthus, Dendronanthus, Tmetothylacus, Macronyx and Motacilla, and depending on the taxonomy followed, Amaurocichla and Madanga) and 66–68 recognised species, of which 32 species in four genera occur in sub- Saharan Africa. The taxonomy of the Motacillidae has been contentious, with variable numbers of genera, species and subspecies proposed and some studies suggesting greater taxonomic diversity than what is currently (five genera and 67 species) recognised. Using one nuclear (Mb) and two mitochondrial (cyt b and CO1) gene regions amplified from DNA extracted from contemporary and museum specimens, we investigated the taxonomic status of 56 of the currently recognised motacillid species and present the most taxonomically complete and expanded phylogeny of this family to date. Our results suggest that the family comprises six clades broadly reflecting continental distributions: sub-Saharan Africa (two clades), the New World (one clade), Palaearctic (one clade), a widespread large-bodied Anthus clade, and a sixth widespread genus, Motacilla. -

North East - Eagle’S Nest & Beyond 7Th April to 20Th April 2022 (14 Days)
India North East - Eagle’s Nest & Beyond 7th April to 20th April 2022 (14 days) Himalayan Cutia by Markus Lilje This exotic birding adventure takes us to many seldom-visited and remote destinations in north-east India. We will have the rare opportunity to search for a host of Asia’s most tantalising and desirable species, including the stunning Temminck’s Tragopan, Bengal Florican, Black-breasted Parrotbill, Ward’s Trogon, exquisite Beautiful Nuthatch, Bugun Liocichla, incredible Grandala, rare and endangered White-winged Duck, and dazzling Fire-tailed Myzornis – to name just some of the many exciting possibilities! This exciting tour takes us to some of Asia’s best birding sites where few birders have ever ventured and we will no doubt encounter many rarely-seen species! RBL India - North-Eastern Itinerary & Extensions 2 THE TOUR AT A GLANCE… THE ITINERARY Day 1 Guwahati to Nameri National Park Day 2 Nameri National Park Day 3 Nameri to Dirang (Sangti Valley) Days 4 to 6 Mandala Phudung and Sela Pass areas Day 7 Dirang to Eagle’s Nest Wildlife Sanctuary Days 8 to 12 Eagle’s Nest Wildlife Sanctuary Day 13 Eagle’s Nest to Tezepur Day 14 Tezepur to Guwahati and depart TOUR ROUTE MAP… RBL India - North-Eastern Itinerary & Extensions 3 THE TOUR IN DETAIL… Day 1: Guwahati to Nameri National Park. The tour will start with a drive from Guwahati to Nameri National Park. This wonderful reserve is contiguous with the Pakhui Wildlife Sanctuary in Arunachal Pradesh, together constituting an area of over 1 000km² (390 mi²) of semi-evergreen forests, moist deciduous forest and narrow strips of grassland along the beautiful crystal-clear, boulder-strewn rivers. -

Scientific Results of the Vernay Lang Kalahari Expedition Birds.Pdf
ANNALE ANNALS VAN DIE OF THE TRANSVAAL MUSEUM VOL. 16 PART I SCIENTIFIC RESULTS OF THE VERNAY- LANG KALAHARI EXPEDITION, MARCH TO SEPTEMBER, 1930 BIRDS By AUSTIN ROBERTS With Plate I HE Vernay-Lang Kalahari Expedition had its inception towards Tthe end of the year 1930, when .:vIr A. S. Vernay arranged with .:vIr Herbert Lang (who was then a guest of the Transvaal .:vIuseum) to combine a sporting and scientific expedition across the unknown central Kalahari to Ngamiland. Mr Vernay had on previous occasions under- taken expeditions for the purpose of securing desiderata in big game animals for museums, and on one occasion Mr Lang had accompanied him and secured much other zoological material for study purposes in Angola. Mr Lang was specially qualified to undertake explorations of this nature from earlier experiences when engaged by the American Museum of Natural History, in which he was Associate Curator of Mammalogy, in collecting specimens in East Africa and the north-eastern Congo. His extraordinary success in the accomplishment of these enterprises had already earned for him a world-wide reputation, and to these successes may be added the present one, as the detailed reports will show when published in due course. Mr Lang has been able to demonstrate that it is not the acquisition of only a few specimens, which become the jealously guarded possessions of a single institution, that makes the suc- cess of an expedition, but tbat by the employment of men who know their business and by their careful collecting of material in such quantity as the particular cases warrant, important facts of value to science can be accumulated. -

Jammu & Kashmir
Biological Forum – An International Journal 11(2): 27-49(2019) ISSN No. (Print): 0975-1130 ISSN No. (Online): 2249-3239 A Preliminary Survey of Bird Communities around Jammu (Jammu & Kashmir) Asha Sohil and Neeraj Sharma* Institute of Mountain Environment, University of Jammu (Corresponding author: Neeraj Sharma) (Received 22 May 2019, Accepted 15 August, 2019) (Published by Research Trend, Website: www.researchtrend.net) ABSTRACT: Landscape heterogeneity strongly influences the bird community structure. With the objectives to understand the richness, assemblages and guild structure of the bird communities in the contagious habitats around Jammu, we carried out the organized avian surveys from January 2017 to December 2017. During this study, 207 species of birds belonging to 63 families and 16 orders were recorded from six contrasting habitats (aquatic and terrestrial). Order Passeriformes (35 families) and family Muscicapidae (17 species) numerically dominated the area. The protected areas and semi disturbed landscapes revealed higher species richness while the moderately disturbed aquatic habitats reflected higher dominance. Based on our observations, 89 birds were found migratory and 10 as globally threatened. The baseline information so generated will prove handy in devising conservation and management plan for birds around Jammu. Keywords: Bird communities, assemblages, guild structure, contrasting habitats, globally threatened, management plan. How to cite this article: Sohil, Asha and Sharma, Neeraj (2019). A preliminary Survey of Bird Communities around Jammu, (Jammu & Kashmir). Biological Forum – An International Journal, 11(2): 27-49. INTRODUCTION includes the work of Pfister (2001), Gupta (2002), Birds are forestanding species of global biodiversity Ahmed and Sahi (2005), Namgail (2005), Kumar and found in every habitat (Olechnowski, 2009) and key Sahi (2006), Wani and Sahi (2007), Aggarwal et al., indicators of ecosystem health and stress (Taper et al., (2008), Kotwal et al., (2009), Namgail and Yom-Tov 1995).