A Genome-Wide Imaging-Based Screening to Identify Genes Involved in Synphilin-1 Inclusion Formation in Saccharomyces Cerevisiae
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Parkin Antibody (C-Term) Purified Rabbit Polyclonal Antibody (Pab) Catalog # AP6402B
10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 Parkin Antibody (C-term) Purified Rabbit Polyclonal Antibody (Pab) Catalog # AP6402B Specification Parkin Antibody (C-term) - Product Information Application IF, WB, IHC-P, FC,E Primary Accession O60260 Other Accession NP_004553 Reactivity Human, Mouse Host Rabbit Clonality Polyclonal Isotype Rabbit Ig Antigen Region 387-417 Parkin Antibody (C-term) - Additional Information Gene ID 5071 Other Names E3 ubiquitin-protein ligase parkin, 632-, Confocal immunofluorescent analysis of Parkinson juvenile disease protein 2, Parkin Antibody (C-term)(Cat#AP6402b) with Parkinson disease protein 2, PARK2, PRKN NCI-H460 cell followed by Alexa Fluor 488-conjugated goat anti-rabbit lgG Target/Specificity (green).Actin filaments have been labeled This Parkin antibody is generated from with Alexa Fluor 555 phalloidin (red).DAPI rabbits immunized with a KLH conjugated was used to stain the cell nuclear (blue). synthetic peptide between 387-417 amino acids from the C-terminal region of human Parkin. Dilution IF~~1:10~50 WB~~1:1000 IHC-P~~1:50~100 FC~~1:10~50 Format Purified polyclonal antibody supplied in PBS with 0.09% (W/V) sodium azide. This antibody is purified through a protein A column, followed by peptide affinity purification. Storage Maintain refrigerated at 2-8°C for up to 2 weeks. For long term storage store at -20°C Park2 Antibody (C-term) (Cat. #AP6402b) in small aliquots to prevent freeze-thaw western blot analysis in K562 cell line lysates cycles. (35ug/lane).This demonstrates the Park2 antibody detected the Park2 protein (arrow). -

Analysis of Gene Expression Data for Gene Ontology
ANALYSIS OF GENE EXPRESSION DATA FOR GENE ONTOLOGY BASED PROTEIN FUNCTION PREDICTION A Thesis Presented to The Graduate Faculty of The University of Akron In Partial Fulfillment of the Requirements for the Degree Master of Science Robert Daniel Macholan May 2011 ANALYSIS OF GENE EXPRESSION DATA FOR GENE ONTOLOGY BASED PROTEIN FUNCTION PREDICTION Robert Daniel Macholan Thesis Approved: Accepted: _______________________________ _______________________________ Advisor Department Chair Dr. Zhong-Hui Duan Dr. Chien-Chung Chan _______________________________ _______________________________ Committee Member Dean of the College Dr. Chien-Chung Chan Dr. Chand K. Midha _______________________________ _______________________________ Committee Member Dean of the Graduate School Dr. Yingcai Xiao Dr. George R. Newkome _______________________________ Date ii ABSTRACT A tremendous increase in genomic data has encouraged biologists to turn to bioinformatics in order to assist in its interpretation and processing. One of the present challenges that need to be overcome in order to understand this data more completely is the development of a reliable method to accurately predict the function of a protein from its genomic information. This study focuses on developing an effective algorithm for protein function prediction. The algorithm is based on proteins that have similar expression patterns. The similarity of the expression data is determined using a novel measure, the slope matrix. The slope matrix introduces a normalized method for the comparison of expression levels throughout a proteome. The algorithm is tested using real microarray gene expression data. Their functions are characterized using gene ontology annotations. The results of the case study indicate the protein function prediction algorithm developed is comparable to the prediction algorithms that are based on the annotations of homologous proteins. -

Dynamic Molecular Linkers of the Genome: the First Decade of SMC Proteins
Downloaded from genesdev.cshlp.org on October 8, 2021 - Published by Cold Spring Harbor Laboratory Press REVIEW Dynamic molecular linkers of the genome: the first decade of SMC proteins Ana Losada1 and Tatsuya Hirano2,3 1Spanish National Cancer Center (CNIO), Madrid E-28029, Spain; 2Cold Spring Harbor Laboratory, Cold Spring Harbor, New York 11724, USA Structural maintenance of chromosomes (SMC) proteins in eukaryotes. The proposed actions of cohesin and con- are chromosomal ATPases, highly conserved from bac- densins offer a plausible, if not complete, explanation for teria to humans, that play fundamental roles in many the sudden appearance of thread-like “substances” (the aspects of higher-order chromosome organization and chromosomes) and their longitudinal splitting during dynamics. In eukaryotes, SMC1 and SMC3 act as the mitosis, first described by Walther Flemming (1882). core of the cohesin complexes that mediate sister chro- Remarkably, SMC proteins are conserved among the matid cohesion, whereas SMC2 and SMC4 function as three phyla of life, indicating that the basic strategy of the core of the condensin complexes that are essential chromosome organization may be evolutionarily con- for chromosome assembly and segregation. Another served from bacteria to humans. An emerging theme is complex containing SMC5 and SMC6 is implicated in that SMC proteins are dynamic molecular linkers of the DNA repair and checkpoint responses. The SMC com- genome that actively fold, tether, and manipulate DNA plexes form unique ring- or V-shaped structures with strands. Their diverse functions range far beyond chro- long coiled-coil arms, and function as ATP-modulated, mosome segregation, and involve nearly all aspects of dynamic molecular linkers of the genome. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

2020 Program Book
PROGRAM BOOK Note that TAGC was cancelled and held online with a different schedule and program. This document serves as a record of the original program designed for the in-person meeting. April 22–26, 2020 Gaylord National Resort & Convention Center Metro Washington, DC TABLE OF CONTENTS About the GSA ........................................................................................................................................................ 3 Conference Organizers ...........................................................................................................................................4 General Information ...............................................................................................................................................7 Mobile App ....................................................................................................................................................7 Registration, Badges, and Pre-ordered T-shirts .............................................................................................7 Oral Presenters: Speaker Ready Room - Camellia 4.......................................................................................7 Poster Sessions and Exhibits - Prince George’s Exhibition Hall ......................................................................7 GSA Central - Booth 520 ................................................................................................................................8 Internet Access ..............................................................................................................................................8 -

Genome-Wide Association Study of Diabetic Kidney Disease Highlights Biology Involved in Glomerular Basement Membrane Collagen
CLINICAL RESEARCH www.jasn.org Genome-Wide Association Study of Diabetic Kidney Disease Highlights Biology Involved in Glomerular Basement Membrane Collagen Rany M. Salem ,1 Jennifer N. Todd,2,3,4 Niina Sandholm ,5,6,7 Joanne B. Cole ,2,3,4 Wei-Min Chen,8 Darrell Andrews,9 Marcus G. Pezzolesi,10 Paul M. McKeigue,11 Linda T. Hiraki,12 Chengxiang Qiu,13 Viji Nair,14 Chen Di Liao,12 Jing Jing Cao,12 Erkka Valo ,5,6,7 Suna Onengut-Gumuscu,8 Adam M. Smiles,15 Stuart J. McGurnaghan,16 Jani K. Haukka,5,6,7 Valma Harjutsalo,5,6,7,17 Eoin P. Brennan,9 Natalie van Zuydam,18,19 Emma Ahlqvist,20 Ross Doyle,9 Tarunveer S. Ahluwalia ,21 Maria Lajer,21 Maria F. Hughes,9 Jihwan Park,13 Jan Skupien,15 Athina Spiliopoulou,11 Andrew Liu,22 Rajasree Menon,14,23 Carine M. Boustany-Kari,24 Hyun M. Kang,23,25 Robert G. Nelson,26 Ronald Klein,27 Barbara E. Klein,27 Kristine E. Lee ,27 Xiaoyu Gao,28 Michael Mauer,29 Silvia Maestroni,30 Maria Luiza Caramori,29 Ian H. de Boer ,31 Rachel G. Miller,32 Jingchuan Guo ,32 Andrew P. Boright,12 David Tregouet,33,34 Beata Gyorgy,33,34 Janet K. Snell-Bergeon,35 David M. Maahs,36 Shelley B. Bull ,37 Angelo J. Canty,38 Colin N.A. Palmer,39 Lars Stechemesser,40 Bernhard Paulweber,40 Raimund Weitgasser,40,41 Jelizaveta Sokolovska,42 Vita Rovıte,43 Valdis Pırags, 42,44 Edita Prakapiene,45 Lina Radzeviciene,46 Rasa Verkauskiene,46 Nicolae Mircea Panduru,6,47 Leif C. -

Familial Cortical Myoclonus Caused by Mutation in NOL3 by Jonathan Foster Rnsseil DISSERTATION Submitted in Partial Satisfaction
Familial Cortical Myoclonus Caused by Mutation in NOL3 by Jonathan Foster Rnsseil DISSERTATION Submitted in partial satisfaction of the requirements for the degree of DOCTOR OF PHILOSOPHY in Biomedical Sciences in the Copyright 2013 by Jonathan Foster Russell ii I dedicate this dissertation to Mom and Dad, for their adamantine love and support iii No man has earned the right to intellectual ambition until he has learned to lay his course by a star which he has never seen—to dig by the divining rod for springs which he may never reach. In saying this, I point to that which will make your study heroic. For I say to you in all sadness of conviction, that to think great thoughts you must be heroes as well as idealists. Only when you have worked alone – when you have felt around you a black gulf of solitude more isolating than that which surrounds the dying man, and in hope and in despair have trusted to your own unshaken will – then only will you have achieved. Thus only can you gain the secret isolated joy of the thinker, who knows that, a hundred years after he is dead and forgotten, men who never heard of him will be moving to the measure of his thought—the subtile rapture of a postponed power, which the world knows not because it has no external trappings, but which to his prophetic vision is more real than that which commands an army. -Oliver Wendell Holmes, Jr. iv ACKNOWLEDGMENTS I am humbled by the efforts of many, many others who were essential for this work. -

Bioinformatic Identification of Genes Suppressing Genome Instability
Bioinformatic identification of genes suppressing PNAS PLUS genome instability Christopher D. Putnama,b, Stephanie R. Allen-Solteroa,c, Sandra L. Martineza, Jason E. Chana,b, Tikvah K. Hayesa,1, and Richard D. Kolodnera,b,c,d,e,2 aLudwig Institute for Cancer Research, Departments of bMedicine and cCellular and Molecular Medicine, dMoores-University of California at San Diego Cancer Center, and eInstitute of Genomic Medicine, University of California School of Medicine at San Diego, La Jolla, CA 92093 Contributed by Richard D. Kolodner, September 28, 2012 (sent for review August 25, 2011) Unbiased forward genetic screens for mutations causing increased in concert to prevent genome rearrangements (reviewed in 12). gross chromosomal rearrangement (GCR) rates in Saccharomyces Modifications of the original GCR assay demonstrated that sup- cerevisiae are hampered by the difficulty in reliably using qualitative pression of GCRs mediated by segmental duplications and Ty GCR assays to detect mutants with small but significantly increased elements involves additional genes and pathways that do not GCR rates. We therefore developed a bioinformatic procedure using suppress single-copy sequence-mediated GCRs (13–15). Inter- genome-wide functional genomics screens to identify and prioritize estingly, homologs of some GCR-suppressing genes and pathways candidate GCR-suppressing genes on the basis of the shared drug suppress the development of cancer in mammals (16). Most of the sensitivity suppression and similar genetic interactions as known genes that suppress GCRs have been identified through a candi- GCR suppressors. The number of known suppressors was increased date gene approach. Some studies have screened collections of from 75 to 110 by testing 87 predicted genes, which identified un- arrayed S. -

Reading Through the Building Blocks of the Genome: Exonic Variation in PD
UNIVERSITY OF CATANIA INTERNATIONAL PhD PROGRAM IN NEUROSCIENCE XXIX Cycle VALENTINA LA COGNATA READING THROUGH THE BUILDING BLOCKS OF THE GENOME: EXONIC VARIATION IN PARKINSON'S DISEASE PhD Thesis Coordinator: Supervisors: Prof. Salvatore Salomone Prof. Velia D’Agata Dr. Sebastiano Cavallaro January 2017 Copyright © V. La Cognata, 2017 All rights reserved. No part of this book may be reproduced, stored in a retrieval system or transmitted in any form or by any means, without prior permission of the author. The copyright of the published papers remains with the publishers. SUMMARY Abstract ...................................................................................................... 5 Chapter 1 .................................................................................................... 7 PARKINSON’S DISEASE ....................................................................................................... 9 HAS EXONIC VARIATION A ROLE IN PD? ............................................................................12 AIMS OF THE PhD WORK ...................................................................................................13 Chapter 2 .................................................................................................. 15 ABSTRACT ........................................................................................................................17 INTRODUCTION ................................................................................................................18 CNVs: A PREVALENT -

Identification of Novel Dna Damage Response Genes Using Functional Genomics
IDENTIFICATION OF NOVEL DNA DAMAGE RESPONSE GENES USING FUNCTIONAL GENOMICS by Michael Chang A thesis submitted in conformity with the requirements for the degree of Doctor of Philosophy Graduate Department of Biochemistry University of Toronto © Copyright by Michael Chang (2005) Identification of novel DNA damage response genes using functional genomics Doctor of Philosophy, 2005; Michael Chang; Department of Biochemistry, University of Toronto ABSTRACT The genetic information required for life is stored within molecules of DNA. This DNA is under constant attack as a result of normal cellular metabolic processes, as well as exposure to genotoxic agents. DNA damage left unrepaired can result in mutations that alter the genetic information encoded within DNA. Cells have consequently evolved complex pathways to combat damage to their DNA. Defects in the cellular response to DNA damage can result in genomic instability, a hallmark of cancer cells. Identifying all the components required for this response remains an important step in fully elucidating the molecular mechanisms involved. I used functional genomic approaches to identify genes required for the DNA damage response in Saccharomyces cerevisiae. I conducted a screen to identify genes required for resistance to a DNA damaging agent, methyl methanesulfonate, and identified several poorly characterized genes that are necessary for proper S phase progression in the presence of DNA damage. Among the genes identified, ESC4/RTT107 has since been shown to be essential for the resumption of DNA replication after DNA damage. Using genome-wide genetic interaction screens to identify genes that are required for viability in the absence of MUS81 and MMS4, two genes required for resistance to DNA damage, I helped identify ELG1, deletion of which causes DNA replication defects, genomic instability, and an inability to properly recover from DNA damage during S phase. -

Subgroup-Specific Structural Variation Across 1,000 Medulloblastoma Genomes
ARTICLE doi:10.1038/nature11327 Subgroup-specific structural variation across 1,000 medulloblastoma genomes A list of authors and their affiliations appears at the end of the paper Medulloblastoma, the most common malignant paediatric brain tumour, is currently treated with nonspecific cytotoxic therapies including surgery, whole-brain radiation, and aggressive chemotherapy. As medulloblastoma exhibits marked intertumoural heterogeneity, with at least four distinct molecular variants, previous attempts to identify targets for therapy have been underpowered because of small samples sizes. Here we report somatic copy number aberrations (SCNAs) in 1,087 unique medulloblastomas. SCNAs are common in medulloblastoma, and are predominantly subgroup-enriched. The most common region of focal copy number gain is a tandem duplication of SNCAIP, a gene associated with Parkinson’s disease, which is exquisitely restricted to Group 4a. Recurrent translocations of PVT1, including PVT1-MYC and PVT1-NDRG1, that arise through chromothripsis are restricted to Group 3. Numerous targetable SCNAs, including recurrent events targeting TGF-b signalling in Group 3, and NF-kB signalling in Group 4, suggest future avenues for rational, targeted therapy. Brain tumours are the most common cause of childhood oncological Twenty-eight regions of recurrent high-level amplification (copy death, and medulloblastoma is the most common malignant paediatric number $ 5) were identified (Fig. 1d and Supplementary Table 7). brain tumour. Current medulloblastoma therapy including surgical The most prevalent amplifications affected members of the MYC resection, whole-brain and spinal cord radiation, and aggressive family with MYCN predominantly amplified in SHH and Group 4, chemotherapy supplemented by bone marrow transplant yields five- MYC in Group 3, and MYCL1 in SHH medulloblastomas. -
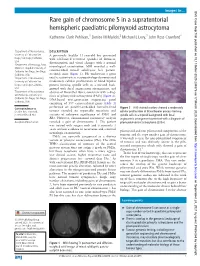
Rare Gain of Chromosome 5 in a Supratentorial Hemispheric
Images in… BMJ Case Rep: first published as 10.1136/bcr-2020-234878 on 17 March 2020. Downloaded from Rare gain of chromosome 5 in a supratentorial hemispheric paediatric pilomyxoid astrocytoma Katherine Clark Pehlivan,1 Denise M Malicki,2 Michael L Levy,3 John Ross Crawford4 1Department of Neurosciences, DESCRIPTION University of California San A previously healthy 11- year- old boy presented Diego, San Diego, California, with self- limited recurrent episodes of dizziness, USA disorientation and visual changes with a normal 2Department of Pathology, Rady neurological examination. MRI revealed a well- Children’s Hospital University of California San Diego, San Diego, circumscribed mixed solid/cystic left parieto- California, USA occipital mass (figure 1). He underwent a gross 3Department of Neurosurgery, total resection where neuropathology demonstrated University of California San moderately cellular proliferation of bland bipolar Diego, San Diego, California, process- forming spindle cells in a myxoid back- USA ground with focal angiocentric arrangement, and 4 Department of Neurosciences absence of Rosenthal fibres, consistent with a diag- and Pediatrics, University of nosis of pilomyxoid astrocytoma (PMA) (figure 2). California San Diego, San Diego, DNA- based next- generation sequencing panel California, USA consisting of 397 cancer- related genes (table 1) performed on paraffin- embedded formalin- fixed Correspondence to Figure 2 H&E- stained sections showed a moderately Dr John Ross Crawford; tumour revealed no reportable mutations and cellular proliferation of bland bipolar process- forming jrcrawford@ ucsd. edu variants of unknown significance of PMS1 and spindle cells in a myxoid background with focal RB1. However, chromosomal microarray analysis angiocentric arrangement consistent with a diagnosis of Accepted 4 March 2020 revealed a gain of chromosome 5.