Poxvirus Exploitation of the Ubiquitin-Proteasome System
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Changes to Virus Taxonomy 2004
Arch Virol (2005) 150: 189–198 DOI 10.1007/s00705-004-0429-1 Changes to virus taxonomy 2004 M. A. Mayo (ICTV Secretary) Scottish Crop Research Institute, Invergowrie, Dundee, U.K. Received July 30, 2004; accepted September 25, 2004 Published online November 10, 2004 c Springer-Verlag 2004 This note presents a compilation of recent changes to virus taxonomy decided by voting by the ICTV membership following recommendations from the ICTV Executive Committee. The changes are presented in the Table as decisions promoted by the Subcommittees of the EC and are grouped according to the major hosts of the viruses involved. These new taxa will be presented in more detail in the 8th ICTV Report scheduled to be published near the end of 2004 (Fauquet et al., 2004). Fauquet, C.M., Mayo, M.A., Maniloff, J., Desselberger, U., and Ball, L.A. (eds) (2004). Virus Taxonomy, VIIIth Report of the ICTV. Elsevier/Academic Press, London, pp. 1258. Recent changes to virus taxonomy Viruses of vertebrates Family Arenaviridae • Designate Cupixi virus as a species in the genus Arenavirus • Designate Bear Canyon virus as a species in the genus Arenavirus • Designate Allpahuayo virus as a species in the genus Arenavirus Family Birnaviridae • Assign Blotched snakehead virus as an unassigned species in family Birnaviridae Family Circoviridae • Create a new genus (Anellovirus) with Torque teno virus as type species Family Coronaviridae • Recognize a new species Severe acute respiratory syndrome coronavirus in the genus Coro- navirus, family Coronaviridae, order Nidovirales -

Kelch Proteins: Emerging Roles in Skeletal Muscle Development and Diseases
Kelch proteins: emerging roles in skeletal muscle development and diseases The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Gupta, Vandana A., and Alan H Beggs. 2014. “Kelch proteins: emerging roles in skeletal muscle development and diseases.” Skeletal Muscle 4 (1): 11. doi:10.1186/2044-5040-4-11. http:// dx.doi.org/10.1186/2044-5040-4-11. Published Version doi:10.1186/2044-5040-4-11 Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:12406733 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA Gupta and Beggs Skeletal Muscle 2014, 4:11 http://www.skeletalmusclejournal.com/content/4/1/11 REVIEW Open Access Kelch proteins: emerging roles in skeletal muscle development and diseases Vandana A Gupta and Alan H Beggs* Abstract Our understanding of genes that cause skeletal muscle disease has increased tremendously over the past three decades. Advances in approaches to genetics and genomics have aided in the identification of new pathogenic mechanisms in rare genetic disorders and have opened up new avenues for therapeutic interventions by identification of new molecular pathways in muscle disease. Recent studies have identified mutations of several Kelch proteins in skeletal muscle disorders. The Kelch superfamily is one of the largest evolutionary conserved gene families. The 66 known family members all possess a Kelch-repeat containing domain and are implicated in diverse biological functions. -

Ankrd9 Is a Metabolically-Controlled Regulator of Impdh2 Abundance and Macro-Assembly
ANKRD9 IS A METABOLICALLY-CONTROLLED REGULATOR OF IMPDH2 ABUNDANCE AND MACRO-ASSEMBLY by Dawn Hayward A dissertation submitted to The Johns Hopkins University in conformity with the requirements of the degree of Doctor of Philosophy Baltimore, Maryland April 2019 ABSTRACT Members of a large family of Ankyrin Repeat Domains proteins (ANKRD) regulate numerous cellular processes by binding and changing properties of specific protein targets. We show that interactions with a target protein and the functional outcomes can be markedly altered by cells’ metabolic state. ANKRD9 facilitates degradation of inosine monophosphate dehydrogenase 2 (IMPDH2), the rate-limiting enzyme in GTP biosynthesis. Under basal conditions ANKRD9 is largely segregated from the cytosolic IMPDH2 by binding to vesicles. Upon nutrient limitation, ANKRD9 loses association with vesicles and assembles with IMPDH2 into rod-like structures, in which IMPDH2 is stable. Inhibition of IMPDH2 with Ribavirin favors ANKRD9 binding to rods. The IMPDH2/ANKRD9 assembly is reversed by guanosine, which restores association of ANKRD9 with vesicles. The conserved Cys109Cys110 motif in ANKRD9 is required for the vesicles-to-rods transition as well as binding and regulation of IMPDH2. ANKRD9 knockdown increases IMPDH2 levels and prevents formation of IMPDH2 rods upon nutrient limitation. Thus, the status of guanosine pools affects the mode of ANKRD9 action towards IMPDH2. Advisor: Dr. Svetlana Lutsenko, Department of Physiology, Johns Hopkins University School of Medicine Second reader: -

Repeat Proteins Challenge the Concept of Structural Domains
844 Biochemical Society Transactions (2015) Volume 43, part 5 Repeat proteins challenge the concept of structural domains Rocıo´ Espada*1, R. Gonzalo Parra*1, Manfred J. Sippl†, Thierry Mora‡, Aleksandra M. Walczak§ and Diego U. Ferreiro*2 *Protein Physiology Lab, Dep de Qu´ımica Biologica, ´ Facultad de Ciencias Exactas y Naturales, UBA-CONICET-IQUIBICEN, Buenos Aires, C1430EGA, Argentina †Center of Applied Molecular Engineering, Division of Bioinformatics, Department of Molecular Biology, University of Salzburg, 5020 Salzburg, Austria ‡Laboratoire de physique statistique, CNRS, UPMC and Ecole normale superieure, ´ 24 rue Lhomond, 75005 Paris, France §Laboratoire de physique theorique, ´ CNRS, UPMC and Ecole normale superieure, ´ 24 rue Lhomond, 75005 Paris, France Abstract Structural domains are believed to be modules within proteins that can fold and function independently. Some proteins show tandem repetitions of apparent modular structure that do not fold independently, but rather co-operate in stabilizing structural forms that comprise several repeat-units. For many natural repeat-proteins, it has been shown that weak energetic links between repeats lead to the breakdown of co-operativity and the appearance of folding sub-domains within an apparently regular repeat array. The quasi-1D architecture of repeat-proteins is crucial in detailing how the local energetic balances can modulate the folding dynamics of these proteins, which can be related to the physiological behaviour of these ubiquitous biological systems. Introduction and between repeats, challenging the concept of structural It was early on noted that many natural proteins typically domain. collapse stretches of amino acid chains into compact units, defining structural domains [1]. These domains typically correlate with biological activities and many modern proteins can be described as composed by novel ‘domain arrange- Definition of the repeat-units ments’ [2]. -

Ectromelia Virus (Mousepox)
technical sheet Ectromelia Virus (Mousepox) Classification the virus its name, ectromelia, or partial amputation of DNA virus, enveloped the limbs and tail. Family Diagnosis Poxviridae Mousepox should be suspected if animals with the above clinical signs are seen in the animal facility, or Affected species there is unexplained widespread mortality in susceptible strains. Serologic diagnosis through MFIA™/ELISA Laboratory mice, wild mice, and other wild rodents or IFA is possible; if animals recover, they produce protective antibodies. Lesions strongly suggestive of Frequency mousepox are noted on necropsy, including splenic Rare in laboratory mice, uncommon in wild mice. fibrosis in recovered animals, and liver, spleen, and skin lesions in ill animals. Histologically, intracytoplasmic Transmission inclusion bodies are seen in skin lesions. PCR of skin Ectromelia virus, which causes the disease mousepox, lesions can be used for confirmation. Vaccination with is transmitted by direct contact or by fomites. Although vaccinia virus as part of an experimental protocol may many routes of infection are possible experimentally, give false positive serology results. exposure to the virus via cutaneous trauma is the natural route of infection. Lesions appear 7-11 days Interference with Research after infection in susceptible strains, and virus is shed Overwhelming mortality (near 100%) in susceptible for 3 weeks. However, virus has been found in scabs strains may have a negative effect on research and feces for as long as 16 weeks post-infection. programs and animal facilities. Ectromelia virus infection Cage-to-cage transmission in mousepox infections is may modify the phagocytic response in resistant primarily through handling of infected mice. -

A Structural Guide to Proteins of the NF-Kb Signaling Module
Downloaded from http://cshperspectives.cshlp.org/ on September 26, 2021 - Published by Cold Spring Harbor Laboratory Press A Structural Guide to Proteins of the NF-kB Signaling Module Tom Huxford1 and Gourisankar Ghosh2 1Department of Chemistry and Biochemistry, San Diego State University, 5500 Campanile Drive, San Diego, California 92182-1030 2Department of Chemistry and Biochemistry, University of California, San Diego, 9500 Gilman Drive, La Jolla, California 92093-0375 Correspondence: [email protected] The prosurvival transcription factor NF-kB specifically binds promoter DNA to activate target gene expression. NF-kB is regulated through interactions with IkB inhibitor proteins. Active proteolysis of these IkB proteins is, in turn, under the control of the IkB kinase complex (IKK). Together, these three molecules form the NF-kB signaling module. Studies aimed at charac- terizing the molecular mechanisms of NF-kB, IkB, and IKK in terms of their three-dimen- sional structures have lead to a greater understanding of this vital transcription factor system. F-kB is a master transcription factor that from the perspective of their three-dimensional Nresponds to diverse cell stimuli by activat- structures. ing the expression of stress response genes. Multiple signals, including cytokines, growth factors, engagement of the T-cell receptor, and NF-kB bacterial and viral products, induce NF-kB Introduction to NF-kB transcriptional activity (Hayden and Ghosh 2008). A point of convergence for the myriad NF-kB was discovered in the laboratory of of NF-kB inducing signals is the IkB kinase David Baltimore as a nuclear activity with bind- complex (IKK). Active IKK in turn controls ing specificity toward a ten-base-pair DNA transcription factor NF-kB by regulating pro- sequence 50-GGGACTTTCC-30 present within teolysis of the IkB inhibitor protein (Fig. -
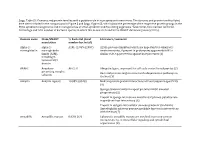
Supp. Table S2: Domains and Protein Families with a Putative Role in Host-Symbiont Interactions
Supp. Table S2: Domains and protein families with a putative role in host-symbiont interactions. The domains and protein families listed here were included in the comparisons in Figure 5 and Supp. Figure S5, which show the percentage of the respective protein groups in the Riftia symbiont metagenome and in metagenomes of other symbiotic and free-living organisms. % bacterial, total number bacterial: Percentage and total number of bacterial species in which this domain is found in the SMART database (January 2019). Domain name Pfam/SMART % bacterial (total Literature/comment annotation number bacterial) Alpha-2- alpha-2- A2M: 42.05% (2057) A2Ms: protease inhibitors which are important for eukaryotic macroglobulin macroglobulin innate immunity, if present in prokaryotes apparently fulfill a family (A2M), similar role, e.g. protection against host proteases (1) including N- terminal MG1 domain ANAPC Anaphase- APC2: 0 Ubiquitin ligase, important for cell cycle control in eukaryotes (2) promoting complex Bacterial proteins might interact with ubiquitination pathways in subunits the host (3) Ankyrin Ankyrin repeats 10.88% (8348) Mediate protein-protein interactions without sequence specificity (4) Sponge symbiont ankyrin-repeat proteins inhibit amoebal phagocytosis (5) Present in sponge microbiome metatranscriptomes, putative role in symbiont-host interactions (6) Present in obligate intracellular amoeba symbiont Candidatus Amoebophilus asiaticus genome, probable function in interactions with the host (7) Armadillo Armadillo repeats 0.83% (67) -

Investigation of Poxvirus Host-Range and Gene Expression in Mammalian Cells
ABSTRACT Title: INVESTIGATION OF POXVIRUS HOST-RANGE AND GENE EXPRESSION IN MAMMALIAN CELLS. Jorge David Méndez Ríos, Doctor of Philosophy, 2014. Directed By: Dr. Bernard Moss, Chief, Laboratory of Viral Diseases, NIAID, NIH; Adjunct Professor Department of Cell Biology and Molecular Genetics, University of Maryland; Members of the Poxviridae family have been known as human pathogens for centuries. Their impact in society included several epidemics that decimated the population. In the last few centuries, Smallpox was of great concern that led to the development of our modern vaccines. The systematic study of Poxvirus host-range and immunogenicity provided the knowledge to translate those observations into practice. After the global vaccination campaign by the World Health Organization, Smallpox was the first infectious disease to be eradicated. Nevertheless, diseases such as Monkeypox, Molluscum contagiosum, new bioterrorist threads, and the use of poxviruses as vaccines or vectors provided the necessity to further understand the host-range from a molecular level. Here, we take advantage of the newly developed technologies such as 454 pyrosequencing and RNA-Seq to address previously unresolved questions for the field. First, we were able to identify the Erytrhomelagia-related poxvirus (ERPV) 25 years after its isolation in Hubei, China. Whole-genome sequencing and bioinformatics identified ERPV as an Ectromelia strain closely related to the Ectromelia Naval strain. Second, by using RNA-Seq, the first MOCV in vivo and in vitro transcriptome was generated. New tools have been developed to support future research and for this human pathogen. Finally, deep-sequencing and comparative genomes of several recombinant MVAs (rMVAs) in conjunction with classical virology allowed us to confirm several genes (O1, F5, C17, F11) association to plaque formation in mammalian cell lines. -

And Γ- Cytoplasmic Actin in Vaccinia Virus Infection
Lights, Camera, Actin: Divergent roles of β- and γ- cytoplasmic actin in vaccinia virus infection NOORUL BISHARA MARZOOK A thesis submitted in fulfillment of requirements for the degree of Doctor of Philosophy FACULTY OF SCIENCE SCHOOL OF MOLECULAR BIOSCIENCE UNIVERSITY OF SYDNEY 2017 i TABLE OF CONTENTS Table of Contents ........................................................................................................... ii Acknowledgements ....................................................................................................... v Declaration ................................................................................................................... vii Abstract ....................................................................................................................... viii List of Figures ................................................................................................................ x List of Publications Arising From This Work.............................................................. xi Abbreviations Used ..................................................................................................... xii Chapter 1: Introduction ............................................................................................... 1 1.1 The Cytoskeleton ............................................................................................................ 2 1.1.1 The Eukaryotic Cytoskeleton ..................................................................................... -

Control of Lymphocytic Choriomeningitis Virus Infection in Granzyme B Deficient Mice
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Virology 305, 1–9 (2003) doi:10.1006/viro.2002.1754 Control of Lymphocytic Choriomeningitis Virus Infection in Granzyme B Deficient Mice Allan J. Zajac,*,† John M. Dye,* and Daniel G. Quinn*,1 *Department of Microbiology and Immunology, Loyola University Chicago, Maywood, Illinois 60153; and †Department of Microbiology, University of Alabama at Birmingham, Birmingham, Alabama 35294 Received July 11, 2001; returned to author for revision October 5, 2001; accepted August 30, 2002 We have investigated whether granzyme B (GzmB)is required for effective cytotoxic T lymphocyte (CTL)mediated control of lymphocytic choriomeningitis virus (LCMV)infection. Clearance of LCMV from tissues of GzmB-deficient (GzmB Ϫ)mice following intraperitoneal infection with LCMV was impaired compared with control mice; however, the virus was ultimately eliminated. The impaired clearance of LCMV in GzmBϪ mice was not due to a deficiency in the generation of LCMV-specific T cells. In addition, CTL from LCMV-infected GzmBϪ mice efficiently lysed virus-infected cells in vitro, but were deficient in their ability to induce rapid DNA fragmentation in target cells. We examined whether the development of protective immunity against intracranial (i.c.)rechallenge with LCMV was compromised in GzmB Ϫ mice. We found that clearance of LCMV from the brain following secondary i.c. infection also was slower in the absence of GzmB; however, the virus was ultimately eliminated and the mice survived. Our data indicate that clearance of LCMV is delayed in the absence of GzmB expression, but that other CTL effector molecules can compensate for the absence of this granule constituent in vivo. -

Appendix 2. Significantly Differentially Regulated Genes in Term Compared with Second Trimester Amniotic Fluid Supernatant
Appendix 2. Significantly Differentially Regulated Genes in Term Compared With Second Trimester Amniotic Fluid Supernatant Fold Change in term vs second trimester Amniotic Affymetrix Duplicate Fluid Probe ID probes Symbol Entrez Gene Name 1019.9 217059_at D MUC7 mucin 7, secreted 424.5 211735_x_at D SFTPC surfactant protein C 416.2 206835_at STATH statherin 363.4 214387_x_at D SFTPC surfactant protein C 295.5 205982_x_at D SFTPC surfactant protein C 288.7 1553454_at RPTN repetin solute carrier family 34 (sodium 251.3 204124_at SLC34A2 phosphate), member 2 238.9 206786_at HTN3 histatin 3 161.5 220191_at GKN1 gastrokine 1 152.7 223678_s_at D SFTPA2 surfactant protein A2 130.9 207430_s_at D MSMB microseminoprotein, beta- 99.0 214199_at SFTPD surfactant protein D major histocompatibility complex, class II, 96.5 210982_s_at D HLA-DRA DR alpha 96.5 221133_s_at D CLDN18 claudin 18 94.4 238222_at GKN2 gastrokine 2 93.7 1557961_s_at D LOC100127983 uncharacterized LOC100127983 93.1 229584_at LRRK2 leucine-rich repeat kinase 2 HOXD cluster antisense RNA 1 (non- 88.6 242042_s_at D HOXD-AS1 protein coding) 86.0 205569_at LAMP3 lysosomal-associated membrane protein 3 85.4 232698_at BPIFB2 BPI fold containing family B, member 2 84.4 205979_at SCGB2A1 secretoglobin, family 2A, member 1 84.3 230469_at RTKN2 rhotekin 2 82.2 204130_at HSD11B2 hydroxysteroid (11-beta) dehydrogenase 2 81.9 222242_s_at KLK5 kallikrein-related peptidase 5 77.0 237281_at AKAP14 A kinase (PRKA) anchor protein 14 76.7 1553602_at MUCL1 mucin-like 1 76.3 216359_at D MUC7 mucin 7, -

Diversity of Large DNA Viruses of Invertebrates ⇑ Trevor Williams A, Max Bergoin B, Monique M
Journal of Invertebrate Pathology 147 (2017) 4–22 Contents lists available at ScienceDirect Journal of Invertebrate Pathology journal homepage: www.elsevier.com/locate/jip Diversity of large DNA viruses of invertebrates ⇑ Trevor Williams a, Max Bergoin b, Monique M. van Oers c, a Instituto de Ecología AC, Xalapa, Veracruz 91070, Mexico b Laboratoire de Pathologie Comparée, Faculté des Sciences, Université Montpellier, Place Eugène Bataillon, 34095 Montpellier, France c Laboratory of Virology, Wageningen University, Droevendaalsesteeg 1, 6708 PB Wageningen, The Netherlands article info abstract Article history: In this review we provide an overview of the diversity of large DNA viruses known to be pathogenic for Received 22 June 2016 invertebrates. We present their taxonomical classification and describe the evolutionary relationships Revised 3 August 2016 among various groups of invertebrate-infecting viruses. We also indicate the relationships of the Accepted 4 August 2016 invertebrate viruses to viruses infecting mammals or other vertebrates. The shared characteristics of Available online 31 August 2016 the viruses within the various families are described, including the structure of the virus particle, genome properties, and gene expression strategies. Finally, we explain the transmission and mode of infection of Keywords: the most important viruses in these families and indicate, which orders of invertebrates are susceptible to Entomopoxvirus these pathogens. Iridovirus Ó Ascovirus 2016 Elsevier Inc. All rights reserved. Nudivirus Hytrosavirus Filamentous viruses of hymenopterans Mollusk-infecting herpesviruses 1. Introduction in the cytoplasm. This group comprises viruses in the families Poxviridae (subfamily Entomopoxvirinae) and Iridoviridae. The Invertebrate DNA viruses span several virus families, some of viruses in the family Ascoviridae are also discussed as part of which also include members that infect vertebrates, whereas other this group as their replication starts in the nucleus, which families are restricted to invertebrates.