Differential Urinary Proteins Between AHSP and Healthy Children Using the DIA Method
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

• Glycolysis • Gluconeogenesis • Glycogen Synthesis
Carbohydrate Metabolism! Wichit Suthammarak – Department of Biochemistry, Faculty of Medicine Siriraj Hospital – Aug 1st and 4th, 2014! • Glycolysis • Gluconeogenesis • Glycogen synthesis • Glycogenolysis • Pentose phosphate pathway • Metabolism of other hexoses Carbohydrate Digestion! Digestive enzymes! Polysaccharides/complex carbohydrates Salivary glands Amylase Pancreas Oligosaccharides/dextrins Dextrinase Membrane-bound Microvilli Brush border Maltose Sucrose Lactose Maltase Sucrase Lactase ‘Disaccharidase’ 2 glucose 1 glucose 1 glucose 1 fructose 1 galactose Lactose Intolerance! Cause & Pathophysiology! Normal lactose digestion Lactose intolerance Lactose Lactose Lactose Glucose Small Intestine Lactase lactase X Galactose Bacteria 1 glucose Large Fermentation 1 galactose Intestine gases, organic acid, Normal stools osmotically Lactase deficiency! active molecules • Primary lactase deficiency: อาการ! genetic defect, การสราง lactase ลด ลงเมออายมากขน, พบมากทสด! ปวดทอง, ถายเหลว, คลนไสอาเจยนภาย • Secondary lactase deficiency: หลงจากรบประทานอาหารทม lactose acquired/transient เชน small bowel เปนปรมาณมาก เชนนม! injury, gastroenteritis, inflammatory bowel disease! Absorption of Hexoses! Site: duodenum! Intestinal lumen Enterocytes Membrane Transporter! Blood SGLT1: sodium-glucose transporter Na+" Na+" •! Presents at the apical membrane ! of enterocytes! SGLT1 Glucose" Glucose" •! Co-transports Na+ and glucose/! Galactose" Galactose" galactose! GLUT2 Fructose" Fructose" GLUT5 GLUT5 •! Transports fructose from the ! intestinal lumen into enterocytes! -

Human Cathepsin A/ Lysosomal Carboxypeptidase a Antibody
Human Cathepsin A/ Lysosomal Carboxypeptidase A Antibody Monoclonal Mouse IgG2A Clone # 179803 Catalog Number: MAB1049 DESCRIPTION Species Reactivity Human Specificity Detects human Cathepsin A/Lysosomal Carboxypeptidase A in direct ELISAs and Western blots. In Western blots, detects the single chain (55 kDa) and heavy chain (32 kDa) forms of recombinant human (rh) Cathepsin A. In Western blots, less than 5% crossreactivity with rhCathepsin B, C, D, E, L, O, S, X and Z is observed and no crossreactivity with the light chain (20 kDa) of rhCathepsin A is observed. Source Monoclonal Mouse IgG2A Clone # 179803 Purification Protein A or G purified from hybridoma culture supernatant Immunogen Mouse myeloma cell line NS0derived recombinant human Cathepsin A/Lysosomal Carboxypeptidase A Ala29Tyr480 (predicted) Accession # P10619 Formulation Lyophilized from a 0.2 μm filtered solution in PBS with Trehalose. See Certificate of Analysis for details. *Small pack size (SP) is supplied either lyophilized or as a 0.2 μm filtered solution in PBS. APPLICATIONS Please Note: Optimal dilutions should be determined by each laboratory for each application. General Protocols are available in the Technical Information section on our website. Recommended Sample Concentration Western Blot 1 µg/mL Recombinant Human Cathepsin A/Lysosomal Carboxypeptidase A (Catalog # 1049SE) Immunoprecipitation 25 µg/mL Conditioned cell culture medium spiked with Recombinant Human Cathepsin A/Lysosomal Carboxypeptidase A (Catalog # 1049SE), see our available Western blot detection antibodies PREPARATION AND STORAGE Reconstitution Reconstitute at 0.5 mg/mL in sterile PBS. Shipping The product is shipped at ambient temperature. Upon receipt, store it immediately at the temperature recommended below. -

Enzymatic Encoding Methods for Efficient Synthesis Of
(19) TZZ__T (11) EP 1 957 644 B1 (12) EUROPEAN PATENT SPECIFICATION (45) Date of publication and mention (51) Int Cl.: of the grant of the patent: C12N 15/10 (2006.01) C12Q 1/68 (2006.01) 01.12.2010 Bulletin 2010/48 C40B 40/06 (2006.01) C40B 50/06 (2006.01) (21) Application number: 06818144.5 (86) International application number: PCT/DK2006/000685 (22) Date of filing: 01.12.2006 (87) International publication number: WO 2007/062664 (07.06.2007 Gazette 2007/23) (54) ENZYMATIC ENCODING METHODS FOR EFFICIENT SYNTHESIS OF LARGE LIBRARIES ENZYMVERMITTELNDE KODIERUNGSMETHODEN FÜR EINE EFFIZIENTE SYNTHESE VON GROSSEN BIBLIOTHEKEN PROCEDES DE CODAGE ENZYMATIQUE DESTINES A LA SYNTHESE EFFICACE DE BIBLIOTHEQUES IMPORTANTES (84) Designated Contracting States: • GOLDBECH, Anne AT BE BG CH CY CZ DE DK EE ES FI FR GB GR DK-2200 Copenhagen N (DK) HU IE IS IT LI LT LU LV MC NL PL PT RO SE SI • DE LEON, Daen SK TR DK-2300 Copenhagen S (DK) Designated Extension States: • KALDOR, Ditte Kievsmose AL BA HR MK RS DK-2880 Bagsvaerd (DK) • SLØK, Frank Abilgaard (30) Priority: 01.12.2005 DK 200501704 DK-3450 Allerød (DK) 02.12.2005 US 741490 P • HUSEMOEN, Birgitte Nystrup DK-2500 Valby (DK) (43) Date of publication of application: • DOLBERG, Johannes 20.08.2008 Bulletin 2008/34 DK-1674 Copenhagen V (DK) • JENSEN, Kim Birkebæk (73) Proprietor: Nuevolution A/S DK-2610 Rødovre (DK) 2100 Copenhagen 0 (DK) • PETERSEN, Lene DK-2100 Copenhagen Ø (DK) (72) Inventors: • NØRREGAARD-MADSEN, Mads • FRANCH, Thomas DK-3460 Birkerød (DK) DK-3070 Snekkersten (DK) • GODSKESEN, -

Diplomarbeit
DIPLOMARBEIT Titel der Diplomarbeit „The influence of free fatty acids on the development of liver inflammation“ Verfasser Mario Kuttke, B.Sc. angestrebter akademischer Grad Magister der Naturwissenschaften (Mag.rer.nat.) Wien, 2012 Studienkennzahl lt. Studienblatt: A 490 Studienrichtung lt. Studienblatt: Diplomstudium Molekulare Biologie Betreuerin / Betreuer: A.o.Univ.-Prof.Dipl.-Ing.Dr. Marcela Hermann Danksagung Zuerst möchte ich mich bei a.o.Univ.-Prof. Dipl.-Ing. Dr. Marcela Hermann für die Betreuung meiner Diplomarbeit bedanken. Besonderer Dank gilt a.o.Univ-Prof. Dr. Bettina Grasl-Kraupp für die fachliche Unterstützung und Betreuung während der praktischen Durchführung der Arbeit. Weiters bedanke ich mich bei Sandra Sagmeister, Therese Böhm, Nora Bintner, Waltraud Schrottmaier, Melanie Pichlbauer, Marzieh Nejabat, Teresa Riegler, Bettina Wingelhofer und Christiane Maier für die ausgezeichnete Zusammenarbeit im Labor und die Unterstützung in allen Lebenslagen. Birgit Mir-Karner, Helga Koudelka und Krystyna Bukowska danke ich für ihre Hilfsbereitschaft und für die kollegiale Zusammenarbeit. Mein größter Dank gilt meinen Eltern, Ursula und Heinz, und meiner Großmutter, Theresia, die mir mein Studium ermöglicht und mich immer unterstützt haben, sowie meinem Bruder, Alex, der in allen Lebenslagen für mich da ist. Table of Contents TABLE OF CONTENTS INTRODUCTION ............................................................................................................................................. 4 HEPATOCELLULAR CARCINOMA (HCC) -

Biochemistry Entry of Fructose and Galactose
Paper : 04 Metabolism of carbohydrates Module : 06 Entry of Fructose and Galactose Dr. Vijaya Khader Dr. MC Varadaraj Principal Investigator Dr.S.K.Khare,Professor IIT Delhi. Paper Coordinator Dr. Ramesh Kothari,Professor UGC-CAS Department of Biosciences Saurashtra University, Rajkot-5, Gujarat-INDIA Dr. S. P. Singh, Professor Content Reviewer UGC-CAS Department of Biosciences Saurashtra University, Rajkot-5, Gujarat-INDIA Dr. Charmy Kothari, Assistant Professor Content Writer Department of Biotechnology Christ College, Affiliated to Saurashtra University, Rajkot-5, Gujarat-INDIA 1 Metabolism of Carbohydrates Biochemistry Entry of Fructose and Galactose Description of Module Subject Name Biochemistry Paper Name 04 Metabolism of Carbohydrates Module Name/Title 06 Entry of Fructose and Galactose 2 Metabolism of Carbohydrates Biochemistry Entry of Fructose and Galactose METABOLISM OF FRUCTOSE Objectives 1. To study the major pathway of fructose metabolism 2. To study specialized pathways of fructose metabolism 3. To study metabolism of galactose 4. To study disorders of galactose metabolism 3 Metabolism of Carbohydrates Biochemistry Entry of Fructose and Galactose Introduction Sucrose disaccharide contains glucose and fructose as monomers. Sucrose can be utilized as a major source of energy. Sucrose includes sugar beets, sugar cane, sorghum, maple sugar pineapple, ripe fruits and honey Corn syrup is recognized as high fructose corn syrup which gives the impression that it is very rich in fructose content but the difference between the fructose content in sucrose and high fructose corn syrup is only 5-10%. HFCS is rich in fructose because the sucrose extracted from the corn syrup is treated with the enzyme that converts some glucose in fructose which makes it more sweet. -

WO 2019/079361 Al 25 April 2019 (25.04.2019) W 1P O PCT
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization I International Bureau (10) International Publication Number (43) International Publication Date WO 2019/079361 Al 25 April 2019 (25.04.2019) W 1P O PCT (51) International Patent Classification: CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, C12Q 1/68 (2018.01) A61P 31/18 (2006.01) DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, C12Q 1/70 (2006.01) HR, HU, ID, IL, IN, IR, IS, JO, JP, KE, KG, KH, KN, KP, KR, KW, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, (21) International Application Number: MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, PCT/US2018/056167 OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, (22) International Filing Date: SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, 16 October 2018 (16. 10.2018) TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (25) Filing Language: English (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (26) Publication Language: English GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, ST, SZ, TZ, (30) Priority Data: UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, 62/573,025 16 October 2017 (16. 10.2017) US TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, EE, ES, FI, FR, GB, GR, HR, HU, ΓΕ , IS, IT, LT, LU, LV, (71) Applicant: MASSACHUSETTS INSTITUTE OF MC, MK, MT, NL, NO, PL, PT, RO, RS, SE, SI, SK, SM, TECHNOLOGY [US/US]; 77 Massachusetts Avenue, TR), OAPI (BF, BJ, CF, CG, CI, CM, GA, GN, GQ, GW, Cambridge, Massachusetts 02139 (US). -
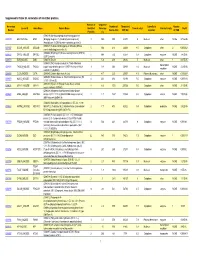
Supporting Table S3 for PDF Maker
Supplemental Table S3. Annotation of identified proteins. Number of Sequence Accession Number of Theoretical Subcellular Number Locus ID Gene Name Protein Name Identified Coverage Theoretical pI Protein Family NSAF Number Amino Acid MW (Da) Location of TMD Peptides (%) (O08539) Myc box-dependent-interacting protein 1 O08539 BIN1_MOUSE BIN1 (Bridging integrator 1) (Amphiphysin-like protein) 3 10.5 588 64470 5 Nucleus other NONE 5.73E-05 (Amphiphysin II) (SH3-domain-containing protein 9) (O08547) Vesicle-trafficking protein SEC22b (SEC22 O08547 SC22B_MOUSE SEC22B 5 30.4 214 24609 8.5 Cytoplasm other 2 0.000262 vesicle-trafficking protein-like 1) (O08553) Dihydropyrimidinase-related protein 2 (DRP-2) O08553 DPYL2_MOUSE DPYSL2 5 19.4 572 62278 6.4 Cytoplasm enzyme NONE 7.85E-05 (ULIP 2 protein) O08579 EMD_MOUSE EMD (O08579) Emerin 1 5.8 259 29436 5 Nucleus other 1 8.67E-05 (O08583) THO complex subunit 4 (Tho4) (RNA and transcription O08583 THOC4_MOUSE THOC4 export factor-binding protein 1) (REF1-I) (Ally of AML-1 1 9.8 254 26809 11.2 Nucleus NONE 2.21E-05 regulator and LEF-1) (Aly/REF) O08585 CLCA_MOUSE CLTA (O08585) Clathrin light chain A (Lca) 2 4.7 235 25557 4.5 Plasma Membrane other NONE 0.000287 (O08600) Endonuclease G, mitochondrial precursor (EC O08600 NUCG_MOUSE ENDOG 4 23.1 294 32191 9.5 Cytoplasm enzyme NONE 0.000134 3.1.30.-) (Endo G) (O08638) Myosin-11 (Myosin heavy chain, smooth O08638 MYH11_MOUSE MYH11 8 6.6 1972 227026 5.5 Cytoplasm other NONE 3.13E-05 muscle isoform) (SMMHC) (O08648) Mitogen-activated protein kinase kinase -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

32-6653: AKR1C1 Human Description Product Info
9853 Pacific Heights Blvd. Suite D. San Diego, CA 92121, USA Tel: 858-263-4982 Email: [email protected] 32-6653: AKR1C1 Human Application : Functional Assay DDH1, DDH, HAKRC, 20-alpha-HSD, DD1/DD2, HBAB, C9, DD1, H-37, MBAB, MGC8954, 2-ALPHA-HSD, Alternative AKR1C1, Aldo-keto reductase family 1 member C1, 20-alpha-hydroxysteroid dehydrogenase, Trans-1,2- Name : dihydrobenzene-1,2-diol dehydrogenase, Indanol dehydrogenase, Dihydrodiol dehydrogenase 1/2, Chlordecone reductase homolog HAKRC, High-affinity hepatic bile acid-binding protein Description Source: Escherichia Coli. Sterile Filtered colorless solution. Aldo-keto reductase family 1 member C1 or AKR1C1 is an enzyme, part of the aldo/keto reductase family that holds over 40 familiar proteins. AKR1C1 promotes the conversion of ketones & aldehydes to their alcohol forms by using cofactors such as NADH & NADPH. AKR1C1 promotes the progesterone reduction to its inactive molecule form 20-alpha-hydroxy-progesterone. AKR1C1 Human Recombinant produced in E.Coli is a single, non-glycosylated polypeptide chain containing 323 amino acids (1-323) and having a molecular mass of 36.7 kDa.AKR1C1 is purified by proprietary chromatographic techniques. Product Info Amount : 2 µg / 10 µg Purification : Greater than 95.0% as determined by SDS-PAGE. The AKR1C1 solution (1mg/ml) contains 20% Glycerol, 0.1M NaCl and 20mM Tris-HCl buffer (pH Content : 8.5). Store at 4°C if entire vial will be used within 2-4 weeks. Store, frozen at -20°C for longer periods of Storage condition : time. For long term storage it is recommended to add a carrier protein (0.1% HSA or BSA).Avoid multiple freeze-thaw cycles. -

Cathepsins and Their Involvement in Immune Responses
Review article: Medical intelligence | Published 20 July 2010, doi:10.4414/smw.2010.13042 Cite this as: Swiss Med Wkly. 2010;140:w13042 Cathepsins and their involvement in immune responses Sébastien Conus, Hans-Uwe Simon Institute of Pharmacology, University of Bern, Bern, Switzerland Correspondence to: cleaving proteases (= caspases) which cleave a wide range Sébastien Conus Ph.D. of cellular substrates [5]. Institute of Pharmacology Besides caspases, cathepsins have recently been shown University of Bern to be associated with cell death regulation [6–12] and vari- Friedbühlstrasse 49 3010 Bern ous other physiological and pathological processes, such as Switzerland maturation of the MHC class II complex, bone remodel- [email protected] ling, keratinocyte differentiation, tumour progression and metastasis, rheumatoid arthritis and osteoarthritis, as well Summary as atherosclerosis [13, 14] (table 1). Thus, cathepsins ap- pear to play a significant role in immune responses. In this The immune system is composed of an enormous variety of review we discuss recent advances addressing the role of cells and molecules that generate a collective and coordin- lysosomal proteases in the diverse aspects of the immune ated response on exposure to foreign antigens, called the response, and also the involvement of cathepsins in the immune response. Within the immune response, endo-lyso- pathogenesis of diseases in which these proteases seem not somal proteases play a key role. In this review we cover to be properly under control. specific roles of cathepsins in innate and adaptive immu- nity, as well as their implication in the pathogenesis of sev- The cathepsin family eral diseases. Lysosomes are membrane-bound organelles which repres- Key words: adaptive and innate immunity; apoptosis; ent the main degradative compartment in eukaryotic cells. -

A Cysteine Protease Inhibitor Blocks SARS-Cov-2 Infection of Human and Monkey Cells
bioRxiv preprint doi: https://doi.org/10.1101/2020.10.23.347534; this version posted October 30, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. A cysteine protease inhibitor blocks SARS-CoV-2 infection of human and monkey cells Drake M. Mellott,1 Chien-Te Tseng,3 Aleksandra Drelich,3 Pavla Fajtová,4,5 Bala C. Chenna,1 Demetrios H. Kostomiris1, Jason Hsu,3 Jiyun Zhu,1 Zane W. Taylor,2,9 Vivian Tat,3 Ardala Katzfuss,1 Linfeng Li,1 Miriam A. Giardini,4 Danielle Skinner,4 Ken Hirata,4 Sungjun Beck4, Aaron F. Carlin,8 Alex E. Clark4, Laura Beretta4, Daniel Maneval6, Felix Frueh,6 Brett L. Hurst,7 Hong Wang,7 Klaudia I. Kocurek,2 Frank M. Raushel,2 Anthony J. O’Donoghue,4 Jair Lage de Siqueira-Neto,4 Thomas D. Meek1.*, and James H. McKerrow#4,* Departments of Biochemistry and Biophysics1 and Chemistry,2 Texas A&M University, 301 Old Main Drive, College Station, Texas 77843, 3Department of Microbiology and Immunology, University of Texas, Medical Branch, 3000 University Boulevard, Galveston, Texas, 77755-1001, 4Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California San Diego, La Jolla, CA, 5Institute of Organic Chemistry and Biochemistry, Academy of Sciences of the Czech Republic, 16610 Prague, Czech Republic, 6Selva Therapeutics, and 7Institute for Antiviral Research, Department of Animal, Dairy, and Veterinary Sciences, 5600 Old Main Hill, Utah State University, Logan, Utah, 84322, 8Department of Medicine, Division of Infectious Diseases and Global Public Health, University of California, San Diego, La Jolla, CA 92037, USA.9Current address: Biological Sciences Division, Pacific Northwest National Laboratory, 902 Battelle Blvd, Richland, WA 99353. -

Coa Protects Against the Deleterious Effects of Caloric Overload in Drosophila Laura Palanker Musselman Washington University School of Medicine in St
Washington University School of Medicine Digital Commons@Becker Open Access Publications 2016 CoA protects against the deleterious effects of caloric overload in Drosophila Laura Palanker Musselman Washington University School of Medicine in St. Louis Jill L. Fink Washington University School of Medicine in St. Louis Thomas J. Baranski Washington University School of Medicine in St. Louis Follow this and additional works at: https://digitalcommons.wustl.edu/open_access_pubs Recommended Citation Musselman, Laura Palanker; Fink, Jill L.; and Baranski, Thomas J., ,"CoA protects against the deleterious effects of caloric overload in Drosophila." Journal of Lipid Research.57,3. 380-387. (2016). https://digitalcommons.wustl.edu/open_access_pubs/5521 This Open Access Publication is brought to you for free and open access by Digital Commons@Becker. It has been accepted for inclusion in Open Access Publications by an authorized administrator of Digital Commons@Becker. For more information, please contact [email protected]. CoA protects against the deleterious effects of caloric overload in Drosophila 1 Laura Palanker Musselman , 2 Jill L. Fink , and Thomas J. Baranski 3 Division of Endocrinology, Metabolism, and Lipid Research, Department of Medicine, Washington University School of Medicine , St. Louis, MO 63110 Abstract We developed a Drosophila model of T2D in in various tissues ( 5–9 ). These lipid mediators induce cel- which high sugar (HS) feeding leads to insulin resistance. In lular stress known as lipid toxicity or “lipotoxicity” ( 4, 10, this model, adipose TG storage is protective against fatty 11 ). Therefore, it is of interest to learn the mechanisms Downloaded from acid toxicity and diabetes. Initial biochemical and gene ex- that enable animals to store excess carbons safely as TG in pression studies suggested that defi ciency in CoA might un- the face of caloric overload.