Sequential Monte Carlo Methods for System Identification
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Prediction Error Methods and Pseudo-Linear Regressions
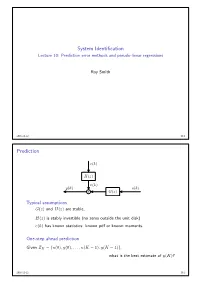
System Identification Lecture 10: Prediction error methods and pseudo-linear regressions Roy Smith 2016-11-22 10.1 Prediction e k p q H z p q v k y k p q u k p q G z p q ` p q Typical assumptions G z and H z are stable, p q p q H z is stably invertible (no zeros outside the unit disk) p q e k has known statistics: known pdf or known moments. p q One-step ahead prediction Given ZK u 0 , y 0 , . , u K 1 , y K 1 , “ t p q p q p ´ q p ´ qu what is the best estimate of y K ? p q 2016-11-22 10.2 Prediction v k e k p q H z p q p q Noise model invertibility Given, v k , k 0,...,K 1, can we determine e k , k 0,...,K 1? p q “ ´ p q “ ´ 8 Inverse filter: Hinv z : e k hinv i v k i p q p q “ i 0 p q p ´ q ÿ“ We also want the inverse filter to be causal and stable: 8 hinv k 0, k 0, and hinv k . p q “ ă k 0 | p q| ă 8 ÿ“ If H z has no zeros for z 1, then, p q | | ě 1 Hinv z . p q “ H z p q 2016-11-22 10.3 Prediction v k e k p q H z p q p q One step ahead prediction Given measurements of v k , k 0,...,K 1, can we predict v K ? p q “ ´ p q Assume that we know H z , how much can we say about v K ? p q p q Assume also that H z is monic (h 0 1). -

10. Prediction Error Methods (PEM)
EE531 - System Identification Jitkomut Songsiri 6. Prediction Error Methods (PEM) • description • optimal prediction • examples • statistical results • computational aspects 6-1 Description idea: determine the model parameter θ such that "(t; θ) = y(t) − y^(tjt − 1; θ) is small y^(tjt − 1; θ) is a prediction of y(t) given the data up to time t − 1 and based on θ general linear predictor: y^(tjt − 1; θ) = N(L; θ)y(t) + M(L; θ)u(t) where M and N must contain one pure delay, i.e., N(0; θ) = 0;M(0; θ) = 0 example: y^(tjt − 1; θ) = 0:5y(t − 1) + 0:1y(t − 2) + 2u(t − 1) Prediction Error Methods (PEM) 6-2 Elements of PEM one has to make the following choices, in order to define the method • model structure: the parametrization of G(L; θ);H(L; θ) and Λ(θ) as a function of θ • predictor: the choice of filters N; M once the model is specified • criterion: define a scalar-valued function of "(t; θ) that will assess the performance of the predictor we commonly consider the optimal mean square predictor the filters N and M are chosen such that the prediction error has small variance Prediction Error Methods (PEM) 6-3 Loss function let N be the number of data points sample covariance matrix: 1 XN R(θ) = "(t; θ)"T (t; θ) N t=1 R(θ) is a positive semidefinite matrix (and typically pdf when N is large) loss function: scalar-valued function defined on positive matrices R f(R(θ)) f must be monotonically increasing, i.e., let X ≻ 0 and for any ∆X ⪰ 0 f(X + ∆X) ≥ f(X) Prediction Error Methods (PEM) 6-4 Example 1 f(X) = tr(WX) where W ≻ 0 is a weighting matrix f(X + ∆X) = tr(WX) + tr(W ∆X) ≥ f(X) (tr(W ∆X) ≥ 0 because if A ⪰ 0;B ⪰ 0, then tr(AB) ≥ 0) Example 2 f(X) = det X f(X + ∆X) − f(X) = det(X1/2(I + X−1/2∆XX−1/2)X1/2) − det X = det X[det(I + X−1/2∆XX−1/2) − 1] " # Yn −1/2 −1/2 = det X (1 + λk(X ∆XX )) − 1 ≥ 0 k=1 −1/2 −1/2 the last inequalty follows from X ∆XX ⪰ 0, so λk ≥ 0 for all k both examples satisfy f(X + ∆X) = f(X) () ∆X = 0 Prediction Error Methods (PEM) 6-5 Procedures in PEM 1. -

5 Identification Using Prediction Error Methods
5 Identification using Prediction Error Methods The previous chapters have shown how the dynamic behaviour of a civil engineering structure can be modelled by a discrete-time ARMAV model or an equivalent stochastic state space realization. The purpose of this chapter is to investigate how an estimate of a p-variate discrete-time model can be obtained from measurements of the response y(tkk). It is assumed that y(t ) is a realization of a Gaussian stochastic process and thus assumed stationary. The stationarity assumption implies that the behaviour of the underlying system can be represented by a time-invariant model. In the following, the general ARMAV(na,nc), for na nc, model will be addressed. In chapter 2, it was shown that this model can be equivalently represented by an m- dimensional state space realization of the innovation form, with m na#p. No matter how the discrete-time model is represented, all adjustable parameters of the model are assembled in a parameter vector , implying that the transfer function description of the discrete-time model becomes y(tk ) H(q, )e(tk ), k 1, 2, ... , N (5.1) T E e(tk )e (tks ) ( ) (s) The dimension of the vectors y(tkk) and e(t ) are p × 1. Since there is no unique way of computing an estimate of (5.1), it is necessary to select an appropriate approach that balances between the required accuracy and computational effort. The approach used in this thesis is known as the Prediction Error Method (PEM), see Ljung [71]. The reason for this choice is that the PEM for Gaussian distributed prediction errors is asymptotic unbiased and efficient. -

Wavelet Analysis for System Identification∗
Wavelet analysis for system identification∗ 大阪教育大学・数理科学 芦野 隆一 (Ryuichi Ashino) Mathematical Sciences, Osaka Kyoiku University 大阪電気通信大学・数理科学研究センター 萬代 武史 (Takeshi Mandai) Research Center for Physics and Mathematics, Osaka Electro-Communication University 大阪教育大学・情報科学 守本 晃 (Akira Morimoto) Information Science, Osaka Kyoiku University Abstract By the Schwartz kernel theorem, to every continuous linear sys- tem there corresponds a unique distribution, called kernel distribution. Formulae using wavelet transform to access time-frequency informa- tion of kernel distributions are deduced. A new wavelet based system identification method for health monitoring systems is proposed as an application of a discretized formula. 1 System Identification A system L illustrated in Figure 1 is an object in which variables of different kinds interact and produce observable signals. The observable signals g that are of interest to us are called outputs. The system is also affected by external stimuli. External signals f that can be manipulated by the observer are called inputs. Others are called disturbances and can be divided into those, denoted by w, that are directly measured, and those, denoted by v, that are only observed through their influence on the output. Mathematically, a system can be regarded as a mapping which relates inputs to outputs. Hereafter, we will use the terminology “system” rather ∗This research was partially supported by the Japanese Ministry of Education, Culture, Sports, Science and Technology, Grant-in-Aid for Scientific Research (C), 15540170 (2003– 2004), and by the Japan Society for the Promotion of Science, Japan-U.S. Cooperative Science Program (2003–2004). 1 Unmeasured Measured disturbance v disturbance w System L Output g Input f Figure 1: System L. -

Localization and System Identification of a Quadcopter AU V
Western Michigan University ScholarWorks at WMU Master's Theses Graduate College 6-2014 Localization and System Identification of a Quadcopter AU V Kenneth Befus Follow this and additional works at: https://scholarworks.wmich.edu/masters_theses Part of the Aeronautical Vehicles Commons, Navigation, Guidance, Control and Dynamics Commons, and the Propulsion and Power Commons Recommended Citation Befus, Kenneth, "Localization and System Identification of a Quadcopter AU V" (2014). Master's Theses. 499. https://scholarworks.wmich.edu/masters_theses/499 This Masters Thesis-Open Access is brought to you for free and open access by the Graduate College at ScholarWorks at WMU. It has been accepted for inclusion in Master's Theses by an authorized administrator of ScholarWorks at WMU. For more information, please contact [email protected]. LOCALIZATION AND SYSTEM IDENTIFICATION OF A QUADCOPTER UAV by Kenneth Befus A thesis submitted to the Graduate College in partial fulfillment of the requirements for the degree of Master of Science in Engineering Department of Mechanical and Aeronautical Engineering Western Michigan University June 2014 Doctoral Committee: Kapseong Ro, Ph.D., Chair Koorosh Naghshineh, Ph.D. James W. Kamman, Ph.D. LOCALIZATION AND SYSTEM IDENTIFICATION OF A QUADCOPTER UAV Kenneth Befus, M.S.E. Western Michigan University, 2014 The research conducted explores the comparison of several trilateration algorithms as they apply to the localization of a quadcopter micro air vehicle (MAV). A localization system is developed employing a network of combined ultrasonic/radio frequency sensors used to wirelessly provide range (distance) measurements defining the location of the quadcopter in 3-dimensional space. A Monte Carlo simulation is conducted using the extrinsic parameters of the localization system to evaluate the adequacy of each trilateration method as it applies to this specific quadcopter application. -

Sequential Monte Carlo Methods for System Identification (A Talk About
Sequential Monte Carlo methods for system identification (a talk about strategies) \The particle filter provides a systematic way of exploring the state space" Thomas Sch¨on Division of Systems and Control Department of Information Technology Uppsala University. Email: [email protected], www: user.it.uu.se/~thosc112 Joint work with: Fredrik Lindsten (University of Cambridge), Johan Dahlin (Link¨opingUniversity), Johan W˚agberg (Uppsala University), Christian A. Naesseth (Link¨opingUniversity), Andreas Svensson (Uppsala University), and Liang Dai (Uppsala University). [email protected] IFAC Symposium on System Identification (SYSID), Beijing, China, October 20, 2015. Introduction A state space model (SSM) consists of a Markov process x ≥ f tgt 1 that is indirectly observed via a measurement process y ≥ , f tgt 1 x x f (x x ; u ); x = a (x ; u ) + v ; t+1 j t ∼ θ t+1 j t t t+1 θ t t θ;t y x g (y x ; u ); y = c (x ; u ) + e ; t j t ∼ θ t j t t t θ t t θ;t x µ (x ); x µ (x ); 1 ∼ θ 1 1 ∼ θ 1 (θ π(θ)): (θ π(θ)): ∼ ∼ Identifying the nonlinear SSM: Find θ based on y y ; y ; : : : ; y (and u ). Hence, the off-line problem. 1:T , f 1 2 T g 1:T One of the key challenges: The states x1:T are unknown. Aim of the talk: Reveal the structure of the system identification problem arising in nonlinear SSMs and highlight where SMC is used. 1 / 14 [email protected] IFAC Symposium on System Identification (SYSID), Beijing, China, October 20, 2015. -

What Is System Identification
What is System identification Using experimental data obtained from input/output relations to model dynamic systems. disturbances input output System Different approaches to system identification depending on model class Linear/Non-linear Parametric/Non-parametric ¾Non-parametric methods try to estimate a generic model. (step responses, impulse responses, frequency responses) ¾Parametric methods estimate parameters in a user-specified model. (transfer functions, state-space matrices) 1 System identification System identification includes the following steps: Experiment design: its purpose is to obtain good experimental data, and it includes the choice of the measured variables and of the character of the input Signals. Selection of model structure: A suitable model structure is chosen using prior knowledge and trial and error. Choice of the criterion to fit: A suitable cost function is chosen, which reflects how well the. model fits the experimental data. Parameter estimation: An optimisation problem is solved to obtain the numerical values of the model parameters. Model validation: The model is tested in order to reveal any inadequacies. 2 Procedure of system identification Experimental design Data collection Data pre-filtering Model structure selection Parameter estimation Model validation Model ok? No Yes 3 Different mathematical models Model description: Transfer functions State-space models Block diagrams (e.g. Simulink) Notation for continuous time models Complex Laplace variable ‘s’ and differential operator ‘p’ ∂x(t) x&()t -

Statistical System Identification and Classification Using Laguerre
Statistical System Identification and Classification Using Laguerre Functions by Pramoda Sachinthana Jayasinghe A Thesis submitted to the Faculty of Graduate Studies of The University of Manitoba in partial fulfilment of the requirements of the degree of Master of Science Department of Statistics University of Manitoba Winnipeg Copyright c 2019 by Pramoda Sachinthana Jayasinghe Abstract This research focuses on developing classification methods to automatically identify partial discharge (PD) signals from multiple sources and developing new system identification techniques to help increase the accuracy of these classifications. We use a Laguerre functional basis to approximate PD signal waveforms, obtained through a lab experiment. To perform the approximation, we considered multiple approaches, such as using objective functional methods and a deterministic method. In the process of signal approximation, we developed methods of selecting a proper scaling factor for Laguerre functions. We evaluated the use of the Laguerre basis expansion coefficients to classify PD signals into their respective sources. Linear discriminant analysis (LDA), quadratic discriminant analysis (QDA) and support vector machines (SVM) were used as classifiers in this analysis. It was observed that these methods can classify partial discharge signals with high accuracies, even when the signals are visually indistinguishable. We also developed two methods for system identification, based on a deterministic approach in the form of a recursive formula and a stochastic method based on a group Lasso model. The aim of system identification was to improve the classification accuracies by removing the effect of the system from the observed PD waveforms. Through numerical evaluations we showed that there are situations where classification after system identification can improve classification accuracies. -

System Identification Via Polynomial Transformation Method
System Identification via Polynomial Transformation Method Prof Pradip Sircar Department of Electrical Engineering Indian Institute of Technology Kanpur Email address: [email protected] Abstract We propose a method based on minimum-variance polynomial approximation to extract system poles from a data set of samples of the impulse response of a linear system. The method is capable of handling the problem under general conditions of sampling and noise characteristics. The superiority of the proposed method is demonstrated by statistical comparison of its performance with the performances of two exiting methods in the special case of uniform sampling. Keywords - Minimum-variance polynomial approximation; System identification; Impulse response; Complex exponential signal Introduction In the all-pole model for linear systems, the characterization of the impulse response by a sum of weighted complex exponential signals, and then estimating the complex frequency- and amplitude- parameters of the signal is equivalent to determining the poles and residues at the poles, respectively, of the system. This particular problem arises in the diversified fields of control, electromagnetics, geophysics, NMR spectroscopy, etc., and is, therefore, of eminent importance. The parameter estimation of complex exponential signals, when the noise corrupted signal samples are provided at non-uniform spacing, has been attempted in [1], [2]. This problem has special significance because non-uniform sampling can be preferred over uniform sampling for various considerations like sampling efficiency, SNR enhancement on the sampled set of signal values, etc. To deal with the non-uniform sampling case, the idea of applying the orthogonal polynomial approximation together with the minimum error-variance criterion to find a closed-form expression for the approximating signal has been introduced in [1], [2]. -

A Novel Logistic-NARX Model As Aclassifier for Dynamic Binary
Neural Comput & Applic DOI 10.1007/s00521-017-2976-x ORIGINAL ARTICLE A novel logistic-NARX model as a classifier for dynamic binary classification Jose Roberto Ayala Solares1 · Hua-Liang Wei1 · Stephen A. Billings1 Received: 12 October 2016 / Accepted: 24 March 2017 © The Natural Computing Applications Forum 2017 Abstract System identification and data-driven modeling outcomes. Furthermore, the NARX approach allows for the techniques have seen ubiquitous applications in the past inclusion of lagged terms and interactions between them in decades. In particular, parametric modeling methodologies a straight forward manner resulting in interpretable mod- such as linear and nonlinear autoregressive with exoge- els where users can identify which input variables play an nous input models (ARX and NARX) and other similar important role individually and/or interactively in the clas- and related model types have been preferably applied to sification process, something that is not achievable using handle diverse data-driven modeling problems due to their other classification techniques like random forests, support easy-to-compute linear-in-the-parameter structure, which vector machines, and k-nearest neighbors. The efficiency of allows the resultant models to be easily interpreted. In the proposed method is tested with five case studies. recent years, several variations of the NARX methodol- ogy have been proposed that improve the performance of Keywords Nonlinear system identification · Dynamic the original algorithm. Nevertheless, in most cases, NARX systems · Binary classification · NARX models · Logistic models are applied to regression problems where all out- regression put variables involve continuous or discrete-time sequences sampled from a continuous process, and little attention has been paid to classification problems where the output sig- 1 Introduction nal is a binary sequence. -

A Bootstrap Subspace Identification Method
Modeling, Identification and Control, Vol. 30, No. 4, 2009, pp. 203{222, ISSN 1890{1328 A Bootstrap Subspace Identification Method: Comparing Methods for Closed Loop Subspace Identification by Monte Carlo Simulations David Di Ruscio Telemark University College, P.O. Box 203, N-3901 Porsgrunn, Norway Abstract A novel promising bootstrap subspace system identification algorithm for both open and closed loop systems is presented. An outline of the SSARX algorithm by Jansson(2003) is given and a modified SSARX algorithm is presented. Some methods which are consistent for closed loop subspace system identification presented in the literature are discussed and compared to a recently published subspace algorithm which works for both open as well as for closed loop data, i.e., the DSR e algorithm as well as the new bootstrap subspace method presented in this paper. Experimental comparisons are performed by Monte Carlo simulations. Keywords: Subspace, Identification, Closed loop, Linear Systems, Kalman filter, Modeling 1. Introduction an optimized version PBSID opt presented. In Chiuso (2007b) the PBSID and PBSID opt methods are fur- Noticing that if the states of a linear dynamic system ther analyzed, and it is shown some asymptotic simi- are known then the problem of finding the model matri- larities with the SSARX and the PBSID algorithms. It ces is a linear regression problem, see e.g., Overschee is also shown that the PBSID opt algorithm is a row and de Moor(1996). Some methods for closed loop weighted version of the SSNEW algorithm in Ljung and subspace identification with state estimation along the McKelvey(1995). Furthermore, Chiuso(2007b) stated lines of the higher order ARX model approach pre- that the PBSID opt algorithm is found to be asymp- sented in Ljung and McKelvey(1995) are presented totically variance efficient on many examples but not in Jansson(2003, 2005) and Chiuso(2007a,b). -

Part 3 – Bayesian System Identification
Learning nonlinear dynamics using sequential Monte Carlo Part 3 { Bayesian system identification Thomas Sch¨on,Uppsala University 2020-03-11 Outline { Part 3 Aim: Show how SMC can be used in identifying nonlinear SSMs using Bayesian approach. Outline: 1. Bayesian inference and the MCMC idea 2. The Metropolis Hastings algorithm 3. Background on Bayesian system identification 4. Using unbiased estimates within Metropolis Hastings 5. Exact approximation { Particle Metropolis Hastings (PMH) 6. Outlook (if there is time) 1/43 Bayesian inference { setup for now Bayesian inference comes down to computing the target distribution π(x). More commonly our interest lies in some integral of the form: Eπ['(x) y1:T ] = '(x)p(x y1:T )dx: j j Z Ex. (nonlinear dynamical systems) Here our interest is often x = θ and π(θ) = p(θ y1:T ) j or x = (x1:T ;θ ) and π(x1:T ;θ ) = p(x1:T ;θ y1:T ). j We keep the development general for now and specialize later. 2/43 How? The two main strategies for the Bayesian inference problem: 1. Variational methods provides an approximation by assuming a certain functional form containing unknown parameters, which are found using optimization, where some distance measure is minimized. 2. Markov chain Monte Carlo (MCMC) works by simulating a Markov chain which is designed in such a way that its stationary distribution coincides with the target distribution. 3/43 MCMC and Metropolis Hastings Toy illustration { AR(1) Let us play the game where you are asked to generate samples from π(x) = x 0; 1=(1 0:82) : N − One realisation from x[t + 1]=0 :8x[t]+ v[t] where v[t] (0; 1).