Comparative Genomics of Amino Acid Tandem Repeats
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The RNA Response to DNA Damage
ÔØ ÅÒÙ×Ö ÔØ The RNA response to DNA damage Luciana E. Giono, Nicol´as Nieto Moreno, Adri´an E. Cambindo Botto, Gwendal Dujardin, Manuel J. Mu˜noz, Alberto R. Kornblihtt PII: S0022-2836(16)00177-7 DOI: doi: 10.1016/j.jmb.2016.03.004 Reference: YJMBI 65022 To appear in: Journal of Molecular Biology Received date: 10 December 2015 Revised date: 1 March 2016 Accepted date: 7 March 2016 Please cite this article as: Giono, L.E., Moreno, N.N., Botto, A.E.C., Dujardin, G., Mu˜noz, M.J. & Kornblihtt, A.R., The RNA response to DNA damage, Journal of Molec- ular Biology (2016), doi: 10.1016/j.jmb.2016.03.004 This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain. ACCEPTED MANUSCRIPT The RNA response to DNA damage Luciana E. Giono1, Nicolás Nieto Moreno1, Adrián E. Cambindo Botto1, Gwendal Dujardin1,2, Manuel J. Muñoz1, and Alberto R. Kornblihtt1* 1Instituto de Fisiología, Biología Molecular y Neurociencias (IFIBYNE-UBA-CONICET) and Departamento de Fisiología, Biología Molecular y Celular, Facultad de Ciencias Exactas y Naturales, Universidad de Buenos Aires, Ciudad Universitaria, Pabellón 2, C1428EHA Buenos Aires, Argentina 2Centre for GenomicACCEPTED Regulation. Dr. -

The Rise and Fall of the Bovine Corpus Luteum
University of Nebraska Medical Center DigitalCommons@UNMC Theses & Dissertations Graduate Studies Spring 5-6-2017 The Rise and Fall of the Bovine Corpus Luteum Heather Talbott University of Nebraska Medical Center Follow this and additional works at: https://digitalcommons.unmc.edu/etd Part of the Biochemistry Commons, Molecular Biology Commons, and the Obstetrics and Gynecology Commons Recommended Citation Talbott, Heather, "The Rise and Fall of the Bovine Corpus Luteum" (2017). Theses & Dissertations. 207. https://digitalcommons.unmc.edu/etd/207 This Dissertation is brought to you for free and open access by the Graduate Studies at DigitalCommons@UNMC. It has been accepted for inclusion in Theses & Dissertations by an authorized administrator of DigitalCommons@UNMC. For more information, please contact [email protected]. THE RISE AND FALL OF THE BOVINE CORPUS LUTEUM by Heather Talbott A DISSERTATION Presented to the Faculty of the University of Nebraska Graduate College in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Biochemistry and Molecular Biology Graduate Program Under the Supervision of Professor John S. Davis University of Nebraska Medical Center Omaha, Nebraska May, 2017 Supervisory Committee: Carol A. Casey, Ph.D. Andrea S. Cupp, Ph.D. Parmender P. Mehta, Ph.D. Justin L. Mott, Ph.D. i ACKNOWLEDGEMENTS This dissertation was supported by the Agriculture and Food Research Initiative from the USDA National Institute of Food and Agriculture (NIFA) Pre-doctoral award; University of Nebraska Medical Center Graduate Student Assistantship; University of Nebraska Medical Center Exceptional Incoming Graduate Student Award; the VA Nebraska-Western Iowa Health Care System Department of Veterans Affairs; and The Olson Center for Women’s Health, Department of Obstetrics and Gynecology, Nebraska Medical Center. -

The Title of the Article
Mechanism-Anchored Profiling Derived from Epigenetic Networks Predicts Outcome in Acute Lymphoblastic Leukemia Xinan Yang, PhD1, Yong Huang, MD1, James L Chen, MD1, Jianming Xie, MSc2, Xiao Sun, PhD2, Yves A Lussier, MD1,3,4§ 1Center for Biomedical Informatics and Section of Genetic Medicine, Department of Medicine, The University of Chicago, Chicago, IL 60637 USA 2State Key Laboratory of Bioelectronics, Southeast University, 210096 Nanjing, P.R.China 3The University of Chicago Cancer Research Center, and The Ludwig Center for Metastasis Research, The University of Chicago, Chicago, IL 60637 USA 4The Institute for Genomics and Systems Biology, and the Computational Institute, The University of Chicago, Chicago, IL 60637 USA §Corresponding author Email addresses: XY: [email protected] YH: [email protected] JC: [email protected] JX: [email protected] XS: [email protected] YL: [email protected] - 1 - Abstract Background Current outcome predictors based on “molecular profiling” rely on gene lists selected without consideration for their molecular mechanisms. This study was designed to demonstrate that we could learn about genes related to a specific mechanism and further use this knowledge to predict outcome in patients – a paradigm shift towards accurate “mechanism-anchored profiling”. We propose a novel algorithm, PGnet, which predicts a tripartite mechanism-anchored network associated to epigenetic regulation consisting of phenotypes, genes and mechanisms. Genes termed as GEMs in this network meet all of the following criteria: (i) they are co-expressed with genes known to be involved in the biological mechanism of interest, (ii) they are also differentially expressed between distinct phenotypes relevant to the study, and (iii) as a biomodule, genes correlate with both the mechanism and the phenotype. -
Integrated Bioinformatics Analysis of Aberrantly-Methylated
Shen et al. BMC Ophthalmology (2020) 20:119 https://doi.org/10.1186/s12886-020-01392-2 RESEARCH ARTICLE Open Access Integrated bioinformatics analysis of aberrantly-methylated differentially- expressed genes and pathways in age- related macular degeneration Yinchen Shen1,2†,MoLi3†, Kun Liu1,2, Xiaoyin Xu1,2, Shaopin Zhu1,2, Ning Wang1,2, Wenke Guo4, Qianqian Zhao4, Ping Lu4, Fudong Yu4 and Xun Xu1,2* Abstract Background: Age-related macular degeneration (AMD) represents the leading cause of visual impairment in the aging population. The goal of this study was to identify aberrantly-methylated, differentially-expressed genes (MDEGs) in AMD and explore the involved pathways via integrated bioinformatics analysis. Methods: Data from expression profile GSE29801 and methylation profile GSE102952 were obtained from the Gene Expression Omnibus database. We analyzed differentially-methylated genes and differentially-expressed genes using R software. Functional enrichment and protein–protein interaction (PPI) network analysis were performed using the R package and Search Tool for the Retrieval of Interacting Genes online database. Hub genes were identified using Cytoscape. Results: In total, 827 and 592 genes showed high and low expression, respectively, in GSE29801; 4117 hyper-methylated genes and 511 hypo-methylated genes were detected in GSE102952. Based on overlap, we categorized 153 genes as hyper-methylated, low-expression genes (Hyper-LGs) and 24 genes as hypo-methylated, high-expression genes (Hypo-HGs). Four Hyper-LGs (CKB, PPP3CA, TGFB2, SOCS2) overlapped with AMD risk genes in the Public Health Genomics and Precision Health Knowledge Base. KEGG pathway enrichment analysis indicated that Hypo-HGs were enriched in the calcium signaling pathway, whereas Hyper-LGs were enriched in sphingolipid metabolism. -

Association of Gene Ontology Categories with Decay Rate for Hepg2 Experiments These Tables Show Details for All Gene Ontology Categories
Supplementary Table 1: Association of Gene Ontology Categories with Decay Rate for HepG2 Experiments These tables show details for all Gene Ontology categories. Inferences for manual classification scheme shown at the bottom. Those categories used in Figure 1A are highlighted in bold. Standard Deviations are shown in parentheses. P-values less than 1E-20 are indicated with a "0". Rate r (hour^-1) Half-life < 2hr. Decay % GO Number Category Name Probe Sets Group Non-Group Distribution p-value In-Group Non-Group Representation p-value GO:0006350 transcription 1523 0.221 (0.009) 0.127 (0.002) FASTER 0 13.1 (0.4) 4.5 (0.1) OVER 0 GO:0006351 transcription, DNA-dependent 1498 0.220 (0.009) 0.127 (0.002) FASTER 0 13.0 (0.4) 4.5 (0.1) OVER 0 GO:0006355 regulation of transcription, DNA-dependent 1163 0.230 (0.011) 0.128 (0.002) FASTER 5.00E-21 14.2 (0.5) 4.6 (0.1) OVER 0 GO:0006366 transcription from Pol II promoter 845 0.225 (0.012) 0.130 (0.002) FASTER 1.88E-14 13.0 (0.5) 4.8 (0.1) OVER 0 GO:0006139 nucleobase, nucleoside, nucleotide and nucleic acid metabolism3004 0.173 (0.006) 0.127 (0.002) FASTER 1.28E-12 8.4 (0.2) 4.5 (0.1) OVER 0 GO:0006357 regulation of transcription from Pol II promoter 487 0.231 (0.016) 0.132 (0.002) FASTER 6.05E-10 13.5 (0.6) 4.9 (0.1) OVER 0 GO:0008283 cell proliferation 625 0.189 (0.014) 0.132 (0.002) FASTER 1.95E-05 10.1 (0.6) 5.0 (0.1) OVER 1.50E-20 GO:0006513 monoubiquitination 36 0.305 (0.049) 0.134 (0.002) FASTER 2.69E-04 25.4 (4.4) 5.1 (0.1) OVER 2.04E-06 GO:0007050 cell cycle arrest 57 0.311 (0.054) 0.133 (0.002) -

Noelia Díaz Blanco
Effects of environmental factors on the gonadal transcriptome of European sea bass (Dicentrarchus labrax), juvenile growth and sex ratios Noelia Díaz Blanco Ph.D. thesis 2014 Submitted in partial fulfillment of the requirements for the Ph.D. degree from the Universitat Pompeu Fabra (UPF). This work has been carried out at the Group of Biology of Reproduction (GBR), at the Department of Renewable Marine Resources of the Institute of Marine Sciences (ICM-CSIC). Thesis supervisor: Dr. Francesc Piferrer Professor d’Investigació Institut de Ciències del Mar (ICM-CSIC) i ii A mis padres A Xavi iii iv Acknowledgements This thesis has been made possible by the support of many people who in one way or another, many times unknowingly, gave me the strength to overcome this "long and winding road". First of all, I would like to thank my supervisor, Dr. Francesc Piferrer, for his patience, guidance and wise advice throughout all this Ph.D. experience. But above all, for the trust he placed on me almost seven years ago when he offered me the opportunity to be part of his team. Thanks also for teaching me how to question always everything, for sharing with me your enthusiasm for science and for giving me the opportunity of learning from you by participating in many projects, collaborations and scientific meetings. I am also thankful to my colleagues (former and present Group of Biology of Reproduction members) for your support and encouragement throughout this journey. To the “exGBRs”, thanks for helping me with my first steps into this world. Working as an undergrad with you Dr. -

Technical Note, Appendix: an Analysis of Blood Processing Methods to Prepare Samples for Genechip® Expression Profiling (Pdf, 1
Appendix 1: Signature genes for different blood cell types. Blood Cell Type Source Probe Set Description Symbol Blood Cell Type Source Probe Set Description Symbol Fraction ID Fraction ID Mono- Lympho- GSK 203547_at CD4 antigen (p55) CD4 Whitney et al. 209813_x_at T cell receptor TRG nuclear cytes gamma locus cells Whitney et al. 209995_s_at T-cell leukemia/ TCL1A Whitney et al. 203104_at colony stimulating CSF1R lymphoma 1A factor 1 receptor, Whitney et al. 210164_at granzyme B GZMB formerly McDonough (granzyme 2, feline sarcoma viral cytotoxic T-lymphocyte- (v-fms) oncogene associated serine homolog esterase 1) Whitney et al. 203290_at major histocompatibility HLA-DQA1 Whitney et al. 210321_at similar to granzyme B CTLA1 complex, class II, (granzyme 2, cytotoxic DQ alpha 1 T-lymphocyte-associated Whitney et al. 203413_at NEL-like 2 (chicken) NELL2 serine esterase 1) Whitney et al. 203828_s_at natural killer cell NK4 (H. sapiens) transcript 4 Whitney et al. 212827_at immunoglobulin heavy IGHM Whitney et al. 203932_at major histocompatibility HLA-DMB constant mu complex, class II, Whitney et al. 212998_x_at major histocompatibility HLA-DQB1 DM beta complex, class II, Whitney et al. 204655_at chemokine (C-C motif) CCL5 DQ beta 1 ligand 5 Whitney et al. 212999_x_at major histocompatibility HLA-DQB Whitney et al. 204661_at CDW52 antigen CDW52 complex, class II, (CAMPATH-1 antigen) DQ beta 1 Whitney et al. 205049_s_at CD79A antigen CD79A Whitney et al. 213193_x_at T cell receptor beta locus TRB (immunoglobulin- Whitney et al. 213425_at Homo sapiens cDNA associated alpha) FLJ11441 fis, clone Whitney et al. 205291_at interleukin 2 receptor, IL2RB HEMBA1001323, beta mRNA sequence Whitney et al. -

Genome-Wide Transcriptional Sequencing Identifies Novel Mutations in Metabolic Genes in Human Hepatocellular Carcinoma DAOUD M
CANCER GENOMICS & PROTEOMICS 11 : 1-12 (2014) Genome-wide Transcriptional Sequencing Identifies Novel Mutations in Metabolic Genes in Human Hepatocellular Carcinoma DAOUD M. MEERZAMAN 1,2 , CHUNHUA YAN 1, QING-RONG CHEN 1, MICHAEL N. EDMONSON 1, CARL F. SCHAEFER 1, ROBERT J. CLIFFORD 2, BARBARA K. DUNN 3, LI DONG 2, RICHARD P. FINNEY 1, CONSTANCE M. CULTRARO 2, YING HU1, ZHIHUI YANG 2, CU V. NGUYEN 1, JENNY M. KELLEY 2, SHUANG CAI 2, HONGEN ZHANG 2, JINGHUI ZHANG 1,4 , REBECCA WILSON 2, LAUREN MESSMER 2, YOUNG-HWA CHUNG 5, JEONG A. KIM 5, NEUNG HWA PARK 6, MYUNG-SOO LYU 6, IL HAN SONG 7, GEORGE KOMATSOULIS 1 and KENNETH H. BUETOW 1,2 1Center for Bioinformatics and Information Technology, National Cancer Institute, Rockville, MD, U.S.A.; 2Laboratory of Population Genetics, National Cancer Institute, National Cancer Institute, Bethesda, MD, U.S.A.; 3Basic Prevention Science Research Group, Division of Cancer Prevention, National Cancer Institute, Bethesda, MD, U.S.A; 4Department of Biotechnology/Computational Biology, St. Jude Children’s Research Hospital, Memphis, TN, U.S.A.; 5Department of Internal Medicine, University of Ulsan College of Medicine, Asan Medical Center, Seoul, Korea; 6Department of Internal Medicine, University of Ulsan College of Medicine, Ulsan University Hospital, Ulsan, Korea; 7Department of Internal Medicine, College of Medicine, Dankook University, Cheon-An, Korea Abstract . We report on next-generation transcriptome Worldwide, liver cancer is the fifth most common cancer and sequencing results of three human hepatocellular carcinoma the third most common cause of cancer-related mortality (1). tumor/tumor-adjacent pairs. -

Molecular Evolutionary Analysis of Plastid Genomes in Nonphotosynthetic Angiosperms and Cancer Cell Lines
The Pennsylvania State University The Graduate School Department or Biology MOLECULAR EVOLUTIONARY ANALYSIS OF PLASTID GENOMES IN NONPHOTOSYNTHETIC ANGIOSPERMS AND CANCER CELL LINES A Dissertation in Biology by Yan Zhang 2012 Yan Zhang Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Dec 2012 The Dissertation of Yan Zhang was reviewed and approved* by the following: Schaeffer, Stephen W. Professor of Biology Chair of Committee Ma, Hong Professor of Biology Altman, Naomi Professor of Statistics dePamphilis, Claude W Professor of Biology Dissertation Adviser Douglas Cavener Professor of Biology Head of Department of Biology *Signatures are on file in the Graduate School iii ABSTRACT This thesis explores the application of evolutionary theory and methods in understanding the plastid genome of nonphotosynthetic parasitic plants and role of mutations in tumor proliferations. We explore plastid genome evolution in parasitic angiosperms lineages that have given up the primary function of plastid genome – photosynthesis. Genome structure, gene contents, and evolutionary dynamics were analyzed and compared in both independent and related parasitic plant lineages. Our studies revealed striking similarities in changes of gene content and evolutionary dynamics with the loss of photosynthetic ability in independent nonphotosynthetic plant lineages. Evolutionary analysis suggests accelerated evolution in the plastid genome of the nonphotosynthetic plants. This thesis also explores the application of phylogenetic and evolutionary analysis in cancer biology. Although cancer has often been likened to Darwinian process, very little application of molecular evolutionary analysis has been seen in cancer biology research. In our study, phylogenetic approaches were used to explore the relationship of several hundred established cancer cell lines based on multiple sequence alignments constructed with variant codons and residues across 494 and 523 genes. -
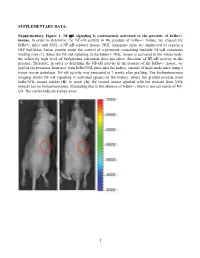
Activation of NF-Κb Signaling Promotes Prostate Cancer Progression in the Mouse and Predicts Poor Progression and Death in Pati
SUPPLEMENTARY DATA: Supplementary Figure 1. NF-B signaling is continuously activated in the prostate of - mouse. In order to determine the NF- '- mouse, we crossed the '- mice with NGL, a NF-B reporter mouse. NGL transgenic mice are engineered to express a GFP/luciferase fusion protein under the control of a promoter containing multiple NF-B consensus binding sites (1). Since the NF-'- NGL mouse is activated in the whole body, the relatively high level of background activation does not allow detection of NF- prostate. Therefore, in order to determine the NF-the '- mouse, we grafted the prostates from 'o the kidney capsule of male nude mice using a tissue rescue technique. NF-B activity was measured at 7 weeks after grafting. The bioluminescence imaging shows NF-B signaling is activated (green) in the kidney, where the grafted prostate from ' use resides (B). In panel (A), the control mouse (grafted with the prostate from NGL mouse) has no bioluminescence, illustrating that in the absence of '-, there is not activation of NF- B. The circles indicate kidney areas. 1 Supplementary Figure 2. NF-B signaling activated in the prostate of Myc/IB bigenic mouse. The prostates from Myc alone (Myc) and bigenic (Myc/IB) mice were harvested at 6 months of age. Activation of NF-B signaling in the prostate was determined by IHC staining of p65-pho antibody. 2 Supplementary Figure 3. Continuous activation of NF-B signaling promotes PCa progression in the Hi-Myc transgenic mouse. The prostates from Myc alone (Myc) and bigeneic (Myc/IB) mice were harvested at 6 months of age. -

Supplementary Table S1
Supplemental Data Table 1A Gene Regulation in PBMCs from normal donors following 18hr IFN-α treatment in vitro Fold Change Post vs Pre- Gene Function treatment Ubiquitin specific protease 18 (USP18) Ubiquitin-dependent protein catabolism 15.1 CD38 antigen (p45) (CD38) Apoptosis 8.8 Interferon-induced protein 44 (IFI44) Antiviral response 7.5 DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 (DDX58) Helicase, Deoxyribonuclease, ubiquitin-protein ligase 7.1 Hect domain and RLD5 (HERC5) Regulation of cyclin dependent protein kinase activity 7.0 2'-5'-oligoadenylate synthetase 2 (OAS2) Immune response, nucleic acid metabolism 6.7 2'-5'-oligoadenylate synthetase-like (OASL) Immune response, nucleic acid metabolism 6.2 Lymphocyte antigen 6 complex, locus E (LY6E) T cell differentiation and activation, antiviral response 5.8 Serine palmitoyltransferase, long chain base subunit 2 (SPTLC2) Unknown 4.6 Likely ortholog of mouse D11lgp2 (LGP2) DNA restriction 4.3 Interferon stimulated gene 20kDa (ISG20) Immune response, proliferation 3.9* Retinoic acid- and interferon-inducible protein (IFIT2) Immune response 3.8 Three prime repair exonuclease 1 (TREX1) DNA repair and replication 3.6 Metallothionein 1H (MT1H) Metal ion binding 3.4 Phospholipid scramblase 1 (PLSCR1) Phospholipid scramblase 3.2* Metallothionein 1 pseudogene 2 (M1P2) Metal ion binding 3.2 Metallothionein 2A (MT2A) Metal ion binding 3.0 SP110 nuclear body protein (SP110) Regulation of transcription, transcription factor 3.0* Tripartite motif-containing 21 (TRIM21) Protein ubiquitination 2.9 -

Mouse Fam76b Knockout Project (CRISPR/Cas9)
https://www.alphaknockout.com Mouse Fam76b Knockout Project (CRISPR/Cas9) Objective: To create a Fam76b knockout Mouse model (C57BL/6J) by CRISPR/Cas-mediated genome engineering. Strategy summary: The Fam76b gene (NCBI Reference Sequence: NM_176836 ; Ensembl: ENSMUSG00000037808 ) is located on Mouse chromosome 9. 10 exons are identified, with the ATG start codon in exon 1 and the TGA stop codon in exon 10 (Transcript: ENSMUST00000059579). Exon 3~8 will be selected as target site. Cas9 and gRNA will be co-injected into fertilized eggs for KO Mouse production. The pups will be genotyped by PCR followed by sequencing analysis. Note: Exon 3 starts from about 15.04% of the coding region. Exon 3~8 covers 66.47% of the coding region. The size of effective KO region: ~7248 bp. The KO region does not have any other known gene. Page 1 of 9 https://www.alphaknockout.com Overview of the Targeting Strategy Wildtype allele 5' gRNA region gRNA region 3' 1 3 4 5 6 7 8 10 Legends Exon of mouse Fam76b Knockout region Page 2 of 9 https://www.alphaknockout.com Overview of the Dot Plot (up) Window size: 15 bp Forward Reverse Complement Sequence 12 Note: The 900 bp section upstream of Exon 3 is aligned with itself to determine if there are tandem repeats. No significant tandem repeat is found in the dot plot matrix. So this region is suitable for PCR screening or sequencing analysis. Overview of the Dot Plot (down) Window size: 15 bp Forward Reverse Complement Sequence 12 Note: The 2000 bp section downstream of Exon 8 is aligned with itself to determine if there are tandem repeats.