Hao Yan CV 2019
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Journal Quartile 2016 & 2017
Journal Impact Journal Impact QUARTILE QUARTILE NO. Full Journal Title ISSN Factor Factor 2016 2017 2016 2017 1CA‐A CANCER JOURNAL FOR CLINICIANS 0007‐9235 187.040 Q1 244.585 Q1 2 NEW ENGLAND JOURNAL OF MEDICINE 0028‐4793 72.406 Q1 79.258 Q1 3 NATURE REVIEWS DRUG DISCOVERY 1474‐1776 57.000 Q1 50.167 Q1 4 CHEMICAL REVIEWS 0009‐2665 47.928 Q1 52.613 Q1 5 LANCET 0140‐6736 47.831 Q1 53.254 Q1 6 NATURE REVIEWS MOLECULAR CELL BIOLOGY 1471‐0072 46.602 Q1 35.612 Q1 7 JAMA‐JOURNAL OF THE AMERICAN MEDICAL ASSOCIATION 0098‐7484 44.405 Q1 47.661 Q1 8 NATURE BIOTECHNOLOGY 1087‐0156 41.667 Q1 35.724 Q1 9 NATURE REVIEWS GENETICS 1471‐0056 40.282 Q1 41.465 Q1 10 NATURE 0028‐0836 40.137 Q1 41.577 Q1 11 NATURE REVIEWS IMMUNOLOGY 1474‐1733 39.932 Q1 41.982 Q1 12 NATURE MATERIALS 1476‐1122 39.737 Q1 39.235 Q1 13 Nature Nanotechnology 1748‐3387 38.986 Q1 37.490 Q1 14 CHEMICAL SOCIETY REVIEWS 0306‐0012 38.618 Q1 40.182 Q1 15 Nature Photonics 1749‐4885 37.852 Q1 32.521 Q1 16 SCIENCE 0036‐8075 37.205 Q1 41.058 Q1 17 NATURE REVIEWS CANCER 1474‐175X 37.147 Q1 42.784 Q1 18 REVIEWS OF MODERN PHYSICS 0034‐6861 36.917 Q1 36.367 Q1 19 LANCET ONCOLOGY 1470‐2045 33.900 Q1 36.418 Q1 20 PROGRESS IN MATERIALS SCIENCE 0079‐6425 31.140 Q1 23.750 Q1 Journal Impact Journal Impact QUARTILE QUARTILE NO. -

Annual Report: CY2018 Chempubsoc Europe
1 Annual Report: CY2018 ChemPubSoc Europe Growth, change, awards, and sponsorship More scientists, more publications, more competition. As in the past, growth continued unabated worldwide in 2018 and is also reflected in the performance metrics of the ChemPubSoc Europe / Wiley-VCH publishing program, which now consists of 16 titles. The most recent product, ChemSystemsChem, was announced in Fall 2018. In 2018, the journals that are co-owned by ChemPubSoc Europe societies published nearly 9,000 articles (excluding contributions to the online publication ChemViews Magazine). Compared to the previous year, manuscript submissions grew by 12%, and the number of published articles rose by approximately 5%. A record number of full-text downloads, >8.9 million, was witnessed for the ChemPubSoc Europe journals, with increases in usage compared to the same time last year; notable among them ca. +20–30% for ChemCatChem, ChemElectroChem, ChemMedChem, and ChemPlusChem; and ca. +70% for ChemistrySelect and ChemPhotoChem. Approximately 3% of articles published in ChemPubSoc Europe's hybrid journals in 2018 were open access: ca. 10% at ChemBioChem and ca. 5% at ChemCatChem, Chemistry—A European Journal, ChemMedChem, and ChemPhysChem, respectively. ChemPubSoc Europe's gold open access journal, ChemistryOpen, experienced a ca. 47% growth in submissions compared to 2017. Generational change After the retirement of Peter Gölitz in November 2017, additional responsibility was taken on by Guido Herrmann (VP, Co-Managing Director Wiley-VCH, since April 1, 2017), Eva Wille (VP, Executive Director Wiley-VCH), Neville Compton (Editor-in-Chief of Angewandte Chemie and publisher for the GDCh journals), Haymo Ross (Editor-in-Chief of Chemistry—A European Journal and publisher of the ChemPubSoc Europe journals), Theresa Kueckmann (Editor-in-Chief of Chemistry—An Asian Journal and publisher for the Asian Chemical Editorial Society (ACES) journals), and Greta Heydenrych (development of new journals). -

Novartis Chemistry Lectureship 20112012
Angewandte. Angewandte News Chemie Novartis Chemistry Lectureship Barcelona, and was appointed professor there in 2011–2012 1986. Giralt is a member of the Editorial Advisory Awarded … Boards of ChemBioChem and Chemistry Open, and The Novartis Chemistry Lectureship is awarded to the International Advisory Boards of Chemistry— scientists in recognition of outstanding contribu- An Asian Journal and ChemMedChem. Giralts tions to organic and computational chemistry, research interests are in the fields of peptide including applications to biology. We congratulate synthesis, molecular recognition, and structure the recipients featured here, as well as Daniel determination, in particular using NMR spectros- Kahne (Harvard University) and Melanie S. San- copy. He has reported on 15N relaxation NMR ford (University of Michigan), who was recently studies of prolyl oligopeptidase in ChemBio- featured in this section.[1] Chem,[4a] and has discussed shuttle-mediated drug Yujiro Hayashi (Tokyo University of Science) delivery to the brain in a Review in Angewandte studied at Tokyo University and was awarded his Chemie.[4b] PhD in 1992 for work supervised by Koichi Tomislav Rovis (Colorado State University, Narasaka. He was a postdoctoral fellow with Fort Collins, USA) studied at the University of Y. Hayashi Elias J. Corey at Harvard University from 1994– Toronto, and earned his PhD in 1998 directed by 1996, and has been full professor at Tokyo Uni- Mark Lautens. From 1998–2000, he was a postdoc- versity of Science since 2006. Hayashi is on the toral fellow with David A. Evans at Harvard Academic Advisory Board of Advanced Synthesis University, and in 2000, he joined the faculty of & Catalysis and the International Advisory Board Colorado State University, where he currently of the European Journal of Organic Chemistry. -

Comparative Bibliometric Analysis of Publications From
COMPARATIVE BIBLIOMETRIC ANALYSIS OF PUBLICATIONS FROM AUSTRALIAN NUCLEAR SCIENCE & TECHNOLOGY ORGANISATION (ANSTO), PAUL SCHERRER INSTITUTE (PSI), KOREA ATOMIC ENERGY RESEARCH INSTITUTE (KAERI) AND, SHANGHAI INSTITUTE OF APPLIED PHYSICS (SINAP) Prepared by: Dr Berenika M. Webster 1 August 2011 Page 1 Contents 1.Executive summary ............................................................................................................ 3 2. Introduction ....................................................................................................................... 5 3. Methodology ..................................................................................................................... 6 Data source and benchmarks ............................................................................................ 6 Analyses ............................................................................................................................ 6 Productivity Measures: ................................................................................................... 6 Recognition & Influence Measures: ................................................................................ 6 Efficiency Measures: ...................................................................................................... 6 Trend Analysis Measures: .............................................................................................. 7 Impact Factor: ............................................................................................................... -

On Behalf of the Regents of the University of California
AMENllMENT TO WILEY ONLINE AGREEMENT Amendment as of January 1, 2019 to the Agreement (the " Agreement " John Wi ley & Sons, Jnc. ("Wiley US"), a New York corporation, and its affiliates, including without limitation, John Wiley & Son of England and Wales, VCH Verla GmbH & o. a , a company mcorporate un er l 1e aws o ("Wiley Germany") and John Wiley & Sons Australia, Ltd., (.. Wiley Australia"), collectively ''Wiley" as successor m interest to I ey Subscription Services, Inc. and The Regents of the University of California, a non-profit academic institution, with its rinci al offices at !he California Di ital Library, University of California Office of the President, SA (the "Customer"). In signing this Amendment, the parties agree that the following terms shall amend and supersede the tenns of said Agreement as follows: I. Paragraph A.2. shall be amended to replace "Licensee" with "Customer''. 2. Paragraph G. I. "Term and Termination" is amended and the tenn of the License is extended to end on December 31, 2019. 3. All applicable Product Appendices are amended to reflect the Licensed Electronic Products for the 201 9 calendar year and the respective pricing for 2019. 5. Paragraph J.12 in the EAL Amendment 2013-15 is removed for 2019 and Lawrence Livermore National Laboratory (LLNL) no longer receives access to the American Geophysical Union (AGU) titles via the California Digital Library license. For the avoidance of doubt, LLNL has perpetual rights to v. I through 2018 of the AGU titles, as stated in the Perpetual Access rights of the Agreement and there are no fees for LLNL's continued access lo the content. -

JCR Science (1º Y 2º Cuartil) 21/09/2017 BIBLIOTECA
JCR Science (1º y 2º cuartil) 21/09/2017 BIBLIOTECA Índice de materias A Agricultural Economics & Acoustic Agricultural Engineering Policy Agriculture, Dairy & Animal Agriculture, Agronomy Science Multidisciplinary Allergy Anatomy & Morphology Andrology Audiology & Speech- Anesthesiology Astronomy & Astrophysics Language Pathology Automation & Control Systems B Biochemical Research Biochemistry & Molecular Behavioral Sciences Methods Biology Biodiversity Conservation Biology Biophysics Biotechnology & Applied Microbiology C Cardiac & Cardiovascular Cell & Tissue Engineering Cell Biology Systems Chemistry, Inorganic & Chemistry, Analytical Chemistry, Applied Nuclear Chemistry, Medicinal Chemistry, Multidisciplinary Chemistry, Organic Computer Science, Artificial Chemistry, Physical Clinical Neurology Intelligence Computer Science, Computer Science, Computer Science, Cybernetics Hardware & Architecture Information Systems Computer Science, Computer Science, Software Computer Science, Theory & Interdisciplinary Engineering Methods Applications Construction & Building Critical Care Medicine Crystallography Technology D Dentistry, Oral Surgery & Dermatology Developmental Biology Medicine E Education, Scientific Ecology Electrochemistry Disciplines Endocrinology & Emergency Medicine Energy & Fuels Metabolism Engineering, Aerospace Engineering, Biomedica Engineering, Chemical Engineering, Electrical & Engineering, Civil Engineering, Environmental Electronic Engineering, Geological Engineering, Industrial Engineering, Manufacturing Engineering, -

Spotlights on Our Sister Journals: Chem. Asian J. 10/2016
On these pages, we feature aselection computer,click on any of the items to of the excellent work that has recently read the full article. Otherwise please been published in our sister journals. see the DOIs for easy online access If you are reading these pages on a through Wiley Online Library. Anion Recognition M. Albrecht,* H. Yi,O.Kçksal, G. Raabe, F. Pan, A. Valkonen, K. Rissanen CF3:AnElectron-Withdrawing Substituent for Aromatic Anion Acceptors?“Side-On” versus“On-Top” Binding of Halides The place to be! Asubtle interplay of electronic effects at the aro- matic system and at aromatic Hatoms in connection with the polar- izability of the anion controlsthe position of anions in respecttoCF3 substituted aromatics. Chem. Eur.J. DOI: 10.1002/chem.201600249 Protein-Protein Interactions A. López, F. Herranz-Trillo, M. Kotev,M.Gairí, V. Guallar,P.Bernadó, O. Millet, T. Tarragó, E. Giralt* Active-Site-DirectedInhibitors of Prolyl Oligopeptidase Abolish Its Conformational Dynamics Aspoke on the wheels:Acombined approach based on NMR and SAXS experiments complemented by MD simulations has shown that active-site-directed inhibitors of prolyl oligopeptidase (POP) abolish the open/closed conformationalequilibrium. Probably,abol- ishing conformational dynamics by inhibitors causes significant alter- ations to molecular recognition events of POP. ChemBioChem DOI: 10.1002/cbic.201600102 Nanocrystals N. Pradhan* Red-Tuned Mn d–d Emission in Doped Semiconductor Nanocrystals Iprefer it in red:Emissiontuned from yellow to red can be ob- tained from Mn2+-doped semiconductor nanocrystals. By changing the environmentofthe Mn dopant in the crystal lattice,the Mn con- centration and the surfaceligands,the ligand-field splitting, and ChemPhysChem therefore also the Mn d–d emission, can be tuned. -
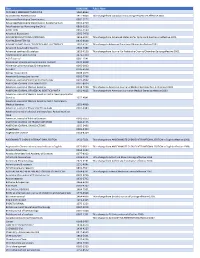
2019 Wiley Title List
Title ISSN/ISBN Public Note ACADEMIC EMERGENCY MEDICINE 1069-6563 ACCOUNTING PERSPECTIVES 1911-382X Title change from Canadian Accounting Perspectives effective 2007. Acta anaesthesiologica Scandinavica 0001-5172 Acta anaesthesiologica Scandinavica. Supplementum 0515-2720 Acta Paediatrica: Nurturing the Child 0803-5253 ADDICTION 0965-2140 Advanced Biosystems 2366-7478 ADVANCED FUNCTIONAL MATERIALS 1616-301X Title change from Advanced Materials for Optics and Electronics effective 2001. ADVANCED MATERIALS 0935-9648 ADVANCED MATERIALS FOR OPTICS AND ELECTRONICS 1057-9257 Title change to Advanced Functional Materials effective 2001. Advanced Sustainable Systems 2366-7486 Advanced synthesis & catalysis 1615-4150 Title change from Journal für Praktische Chemie/Chemiker-Zeitung effective 2001. AEM Education and Training 2472-5390 AIChE Journal 0001-1541 Alcoholism: clinical and experimental research 0145-6008 Alimentary pharmacology & therapeutics 0269-2813 ALLERGY 0105-4538 Allergy. Supplement 0108-1675 American Business Law Journal 0002-7766 American Journal of Community Psychology 0091-0562 AMERICAN JOURNAL OF HEMATOLOGY 0361-8609 American Journal of Medical Genetics 0148-7299 Title change to American Journal of Medical Genetics Part A effeective 2003. AMERICAN JOURNAL OF MEDICAL GENETICS PART A 1552-4825 Title change from American Journal of Medical Genetics effective 2003. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics 1552-4841 American Journal of Medical Genetics Part C: Seminars in Medical Genetics 1552-4868 American Journal of Physical Anthropology 0002-9483 American journal of physical anthropology. Annual meeting issue American Journal of Political Science 0092-5853 AMERICAN JOURNAL OF TRANSPLANTATION 1600-6135 AMERICAN JOURNAL ON ADDICTIONS 1055-0496 Anaesthesia 0003-2409 Angewandte Chemie 0044-8249 ANGEWANDTE CHEMIE INTERNATIONAL EDITION 1433-7851 Title change from ANGEWANDTE CHEMIE INTERNATIONAL EDITION in English effective 1998. -

Jianfeng Cai CV.Pdf
Jianfeng Cai [email protected] USF Preeminent Professor Department of Chemistry University of South Florida Tampa, FL 33620 (Office) 813-974-9506 EDUCATION Postdoctoral Associate, Bioorganic Chemistry, Yale University, 2007-2009 Advisor: Professor Andrew D. Hamilton PhD, Bioorganic Chemistry, Washington University in St. Louis, 2002-2006 Advisor: Professor John-Stephen Taylor Thesis Title: Design and Synthesis of Nucleic Acid Templated and Targeted Drugs and Probes MS, Nanjing University, China, 2000 BS, Nanjing University, China, 1997 POSITIONS AND EMPLOYMENT 2007-2009 Postdoctoral Associate, Yale University, New Haven, CT 2009-2015 Assistant Professor, University of South Florida, Tampa, FL 2015-2018 Associate Professor, University of South Florida, Tampa, FL 2018-Present Professor, University of South Florida, Tampa, FL 2020-Present USF Preeminent Professor, University of South Florida, Tampa, FL 2009-Present Member, Drug Discovery Program, Moffitt Cancer Center, Tampa, FL 2019-Present Director, Center for Molecular Diversity in Drug Design, Discovery and Development (CMD5) AWARDS AND RECOGNITIONS 2021 Outstanding Faculty Award, USF 2020 Outstanding Graduate Faculty Mentor Award, USF 2020 USF Faculty Outstanding Research Achievement Award 2020 USF Preeminent Professor 2020 Member, National Academy of Inventors, USF Chapter 2020 Fellow of Royal Society of Chemistry (FRSC) 2018 USF Faculty Outstanding Research Achievement Award 2015-2017 Outstanding reviewer, Journal of Medicinal Chemistry 2015 USF Faculty Outstanding Research Achievement Award 2015 Biomatik Distinguished Junior Faculty Award, the Chinese-American Chemistry & Chemical Biology Professors Association (CAPA) 2014 Excellence in reviewing, European Journal of Medicinal Chemistry 2014 NSF Career Award 2014 ChemComm Emerging Investigator 2012 New Investigator award, Florida Bankhead Coley Cancer Research Program 2011 Ralph E. -

List of SCIE Journals
REUTERS/Morteza Nikoubazl SOURCE PUBLICATION LIST FOR WEB OF SCIENCE® SCIENCE CITATION INDEX EXPANDEDTM 2014 AUGUST WEB OF SCIENCE SCIE JOURNAL LIST TITLE PUBLISHER ISSN E_ISSN COUNTRY LANGUAGE 4OR-A Quarterly Journal of Operations Research SPRINGER HEIDELBERG 1619-4500 1614-2411 GERMANY English AAPG BULLETIN AMER ASSOC PETROLEUM GEOLOGIST 0149-1423 1558-9153 UNITED STATES English AAPS Journal SPRINGER 1550-7416 1550-7416 UNITED STATES English AAPS PHARMSCITECH SPRINGER 1530-9932 1530-9932 UNITED STATES English AATCC REVIEW AMER ASSOC TEXTILE CHEMISTS COLORISTS 1532-8813 UNITED STATES English ABDOMINAL IMAGING SPRINGER 0942-8925 1432-0509 UNITED STATES English ABHANDLUNGEN AUS DEM MATHEMATISCHEN SEMINAR DER UNIVERSITAT HAMBURG SPRINGER HEIDELBERG 0025-5858 1865-8784 GERMANY German Abstract and Applied Analysis HINDAWI PUBLISHING CORPORATION 1085-3375 1687-0409 UNITED STATES English ABSTRACTS OF PAPERS OF THE AMERICAN CHEMICAL SOCIETY AMER CHEMICAL SOC 0065-7727 UNITED STATES English ACADEMIC EMERGENCY MEDICINE WILEY-BLACKWELL 1069-6563 1553-2712 UNITED STATES English ACADEMIC MEDICINE LIPPINCOTT WILLIAMS & WILKINS 1040-2446 1938-808X UNITED STATES English Academic Pediatrics ELSEVIER SCIENCE INC 1876-2859 1876-2867 UNITED STATES English ACADEMIC RADIOLOGY ELSEVIER SCIENCE INC 1076-6332 1878-4046 UNITED STATES English ACAROLOGIA ACAROLOGIA-UNIVERSITE PAUL VALERY 0044-586X 2107-7207 FRANCE Multi-Language Accountability in Research-Policies and Quality Assurance TAYLOR & FRANCIS LTD 0898-9621 1545-5815 UNITED STATES English ACCOUNTS OF CHEMICAL -

Aqueous Electrolytes for High-Voltage Batteries and Supercapacitors
Thèse n° 8287 Aqueous electrolytes for high-voltage batteries and supercapacitors Présentée le 23 juillet 2020 à la Faculté des sciences et techniques de l’ingénieur Unité de rattachement pour scientifiques IMX Programme doctoral en science et génie des matériaux pour l’obtention du grade de Docteur ès Sciences par David REBER Acceptée sur proposition du jury Prof. P. Hoffmann, président du jury Prof. F. Nüesch, Dr C. Battaglia, directeurs de thèse Dr K. Xu, rapporteur Prof. P. Broekmann, rapporteur Prof. S. Haussener, rapporteuse 2020 Abstract Lithium-ion batteries are widely implemented as energy storage devices due to their high energy density and low cost. They enabled modern portable electronics and electric vehicles, and are key to manage the integration of intermittent renewable electricity sources, such as solar and wind, into the power grid and decentralized microgrids. The use of flammable or- ganic electrolytes however raises safety concerns, particularly for large-scale stationary bat- teries. Furthermore, supply chain risks and the use of critical raw materials are a major chal- lenge for the ongoing battery revolution. Aqueous electrolytes offer promising properties, as they are inherently non-flammable and potentially more cost effective. Combined with electrode materials based on abundant raw materials, aqueous batteries are among the most promising candidates to replace current battery chemistries relying on critical cobalt and flammable organic electrolytes. However, due to the narrow electrochemical stability window of water (practically ca. 1.5 V), aqueous batteries have not yet met the energy density to be serious competitors to current lithium- ion battery technology. The goal of this thesis was to develop aqueous electrolytes with enhanced electrochem- ical stability, thus enabling new high-voltage cell chemistries. -

Chemistry and Biochemistry Authored Faculty Publications
University of Arkansas, Fayetteville ScholarWorks@UARK University Libraries Faculty Publications and Presentations University Libraries 7-30-2020 Chemistry and Biochemistry Authored Faculty Publications: Snapshot of the Ranking of Journals in which they published based on data from Journal Citation Reports (JCR) Lutishoor Salisbury University of Arkansas, Fayetteville, [email protected] Yang Tian University of Arkansas, Fayetteville Jeremy Smith University of Arkansas, Fayetteville Follow this and additional works at: https://scholarworks.uark.edu/libpub Part of the Cataloging and Metadata Commons, Collection Development and Management Commons, Materials Chemistry Commons, and the Physical Chemistry Commons Citation Salisbury, L., Tian, Y., & Smith, J. (2020). Chemistry and Biochemistry Authored Faculty Publications: Snapshot of the Ranking of Journals in which they published based on data from Journal Citation Reports (JCR). University Libraries Faculty Publications and Presentations. Retrieved from https://scholarworks.uark.edu/libpub/38 This Report is brought to you for free and open access by the University Libraries at ScholarWorks@UARK. It has been accepted for inclusion in University Libraries Faculty Publications and Presentations by an authorized administrator of ScholarWorks@UARK. For more information, please contact [email protected]. University of Arkansas Libraries Chemistry and Biochemistry Library Chemistry and Biochemistry Authored Faculty Publications: Snapshot of the Ranking of Journals in which they published based on data from Journal Citation Reports (JCR). (https://go.openathens.net/redirector/uark.edu?url=https%3A%2F%2Fjcr.clarivate.com This report provides a snapshot of the ranking of journals for CHBC faculty authored papers for the period: 2015-2019. JCR is a database produced by Clarivate Analytics that provides impact factors and rankings of over 9000 journals.