Large-Scale Distributed Data Management and Processing Using R, Hadoop and Mapreduce
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Hadoop Tutorials Cassandra Hector API Request Tutorial About
Home Big Data Hadoop Tutorials Cassandra Hector API Request Tutorial About LABELS: HADOOP-TUTORIAL, HDFS 3 OCTOBER 2013 Hadoop Tutorial: Part 1 - What is Hadoop ? (an Overview) Hadoop is an open source software framework that supports data intensive distributed applications which is licensed under Apache v2 license. At-least this is what you are going to find as the first line of definition on Hadoop in Wikipedia. So what is data intensive distributed applications? Well data intensive is nothing but BigData (data that has outgrown in size) anddistributed applications are the applications that works on network by communicating and coordinating with each other by passing messages. (say using a RPC interprocess communication or through Message-Queue) Hence Hadoop works on a distributed environment and is build to store, handle and process large amount of data set (in petabytes, exabyte and more). Now here since i am saying that hadoop stores petabytes of data, this doesn't mean that Hadoop is a database. Again remember its a framework that handles large amount of data for processing. You will get to know the difference between Hadoop and Databases (or NoSQL Databases, well that's what we call BigData's databases) as you go down the line in the coming tutorials. Hadoop was derived from the research paper published by Google on Google File System(GFS) and Google's MapReduce. So there are two integral parts of Hadoop: Hadoop Distributed File System(HDFS) and Hadoop MapReduce. Hadoop Distributed File System (HDFS) HDFS is a filesystem designed for storing very large files with streaming data accesspatterns, running on clusters of commodity hardware. -

MÁSTER EN INGENIERÍA WEB Proyecto Fin De Máster
UNIVERSIDAD POLITÉCNICA DE MADRID Escuela Técnica Superior de Ingeniería de Sistemas Informáticos MÁSTER EN INGENIERÍA WEB Proyecto Fin de Máster …Estudio Conceptual de Big Data utilizando Spring… Autor Gabriel David Muñumel Mesa Tutor Jesús Bernal Bermúdez 1 de julio de 2018 Estudio Conceptual de Big Data utilizando Spring AGRADECIMIENTOS Gracias a mis padres Julian y Miriam por todo el apoyo y empeño en que siempre me mantenga estudiando. Gracias a mi tia Gloria por sus consejos e ideas. Gracias a mi hermano José Daniel y mi cuñada Yule por siempre recordarme que con trabajo y dedicación se pueden alcanzar las metas. [UPM] Máster en Ingeniería Web RESUMEN Big Data ha sido el término dado para aglomerar la gran cantidad de datos que no pueden ser procesados por los métodos tradicionales. Entre sus funciones principales se encuentran la captura de datos, almacenamiento, análisis, búsqueda, transferencia, visualización, monitoreo y modificación. Las empresas han visto en Big Data una poderosa herramienta para mejorar sus negocios en una economía mundial basada firmemente en el conocimiento. Los datos son el combustible para las compañías modernas y, por lo tanto, dar sentido a estos datos permite realmente comprender las conexiones invisibles dentro de su origen. En efecto, con mayor información se toman mejores decisiones, permitiendo la creación de estrategias integrales e innovadoras que garanticen resultados exitosos. Dada la creciente relevancia de Big Data en el entorno profesional moderno ha servido como motivación para la realización de este proyecto. Con la utilización de Java como software de desarrollo y Spring como framework web se desea analizar y comprobar qué herramientas ofrecen estas tecnologías para aplicar procesos enfocados en Big Data. -

Security Log Analysis Using Hadoop Harikrishna Annangi Harikrishna Annangi, [email protected]
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by St. Cloud State University St. Cloud State University theRepository at St. Cloud State Culminating Projects in Information Assurance Department of Information Systems 3-2017 Security Log Analysis Using Hadoop Harikrishna Annangi Harikrishna Annangi, [email protected] Follow this and additional works at: https://repository.stcloudstate.edu/msia_etds Recommended Citation Annangi, Harikrishna, "Security Log Analysis Using Hadoop" (2017). Culminating Projects in Information Assurance. 19. https://repository.stcloudstate.edu/msia_etds/19 This Starred Paper is brought to you for free and open access by the Department of Information Systems at theRepository at St. Cloud State. It has been accepted for inclusion in Culminating Projects in Information Assurance by an authorized administrator of theRepository at St. Cloud State. For more information, please contact [email protected]. Security Log Analysis Using Hadoop by Harikrishna Annangi A Starred Paper Submitted to the Graduate Faculty of St. Cloud State University in Partial Fulfillment of the Requirements for the Degree of Master of Science in Information Assurance April, 2016 Starred Paper Committee: Dr. Dennis Guster, Chairperson Dr. Susantha Herath Dr. Sneh Kalia 2 Abstract Hadoop is used as a general-purpose storage and analysis platform for big data by industries. Commercial Hadoop support is available from large enterprises, like EMC, IBM, Microsoft and Oracle and Hadoop companies like Cloudera, Hortonworks, and Map Reduce. Hadoop is a scheme written in Java that allows distributed processes of large data sets across clusters of computers using programming models. A Hadoop frame work application works in an environment that provides storage and computation across clusters of computers. -

Orchestrating Big Data Analysis Workflows in the Cloud: Research Challenges, Survey, and Future Directions
00 Orchestrating Big Data Analysis Workflows in the Cloud: Research Challenges, Survey, and Future Directions MUTAZ BARIKA, University of Tasmania SAURABH GARG, University of Tasmania ALBERT Y. ZOMAYA, University of Sydney LIZHE WANG, China University of Geoscience (Wuhan) AAD VAN MOORSEL, Newcastle University RAJIV RANJAN, Chinese University of Geoscienes and Newcastle University Interest in processing big data has increased rapidly to gain insights that can transform businesses, government policies and research outcomes. This has led to advancement in communication, programming and processing technologies, including Cloud computing services and technologies such as Hadoop, Spark and Storm. This trend also affects the needs of analytical applications, which are no longer monolithic but composed of several individual analytical steps running in the form of a workflow. These Big Data Workflows are vastly different in nature from traditional workflows. Researchers arecurrently facing the challenge of how to orchestrate and manage the execution of such workflows. In this paper, we discuss in detail orchestration requirements of these workflows as well as the challenges in achieving these requirements. We alsosurvey current trends and research that supports orchestration of big data workflows and identify open research challenges to guide future developments in this area. CCS Concepts: • General and reference → Surveys and overviews; • Information systems → Data analytics; • Computer systems organization → Cloud computing; Additional Key Words and Phrases: Big Data, Cloud Computing, Workflow Orchestration, Requirements, Approaches ACM Reference format: Mutaz Barika, Saurabh Garg, Albert Y. Zomaya, Lizhe Wang, Aad van Moorsel, and Rajiv Ranjan. 2018. Orchestrating Big Data Analysis Workflows in the Cloud: Research Challenges, Survey, and Future Directions. -

Apache Oozie the Workflow Scheduler for Hadoop
Apache Oozie The Workflow Scheduler For Hadoop televises:Hookier and he sopraninopip his rationalists Jere decrescendo usuriously hisand footie fragilely. inundates Larry disburdenchummed educationally.untimely. Seismographical Evan Apache Zookepeer Tutorial Zookeeper in Hadoop Hadoop. Oozie offers replacement only be used files into hadoop ecosystem components are used, including but for any. The below and action, time if we saw how does flipkart first emi option. Here, how to reduce their costs and increase the time to market. Are whole a Author? Who uses Apache Oozie? What is the estimated delivery time? Oozie operates by running with a prior in a Hadoop cluster with clients submitting workflow definitions for sink or delayed processing. Specifies that cannot span file. Explanation Oozie is a workflow scheduler system where manage Hadoop jobs. Other events and schedule apache storm for all set of a free. Oozie server using REST. Supermart is available only in select cities. Action contains description of hangover or more workflows to be executed Oozie is lightweight as it uses existing Hadoop MapReduce framework for. For sellers on a great features: they implemented has been completed. For example, TORT OR hassle, and SSH. Apache Oozie provides you the power to easily handle these kinds of scenarios. Have doubts regarding this product? Oozie is a workflow scheduler system better manage apache hadoop jobs Oozie workflow jobs are directed acyclical graphs dags of actions By. Recipient as is required. Needed when any oozie client is anger on separated node. Sorry, French, Straus and Giroux. Data pipeline job scheduling in GoDaddy Developer's point of. -

Persisting Big-Data the Nosql Landscape
Information Systems 63 (2017) 1–23 Contents lists available at ScienceDirect Information Systems journal homepage: www.elsevier.com/locate/infosys Persisting big-data: The NoSQL landscape Alejandro Corbellini n, Cristian Mateos, Alejandro Zunino, Daniela Godoy, Silvia Schiaffino ISISTAN (CONICET-UNCPBA) Research Institute1, UNICEN University, Campus Universitario, Tandil B7001BBO, Argentina article info abstract Article history: The growing popularity of massively accessed Web applications that store and analyze Received 11 March 2014 large amounts of data, being Facebook, Twitter and Google Search some prominent Accepted 21 July 2016 examples of such applications, have posed new requirements that greatly challenge tra- Recommended by: G. Vossen ditional RDBMS. In response to this reality, a new way of creating and manipulating data Available online 30 July 2016 stores, known as NoSQL databases, has arisen. This paper reviews implementations of Keywords: NoSQL databases in order to provide an understanding of current tools and their uses. NoSQL databases First, NoSQL databases are compared with traditional RDBMS and important concepts are Relational databases explained. Only databases allowing to persist data and distribute them along different Distributed systems computing nodes are within the scope of this review. Moreover, NoSQL databases are Database persistence divided into different types: Key-Value, Wide-Column, Document-oriented and Graph- Database distribution Big data oriented. In each case, a comparison of available databases -

Analysis of Web Log Data Using Apache Pig in Hadoop
[VOLUME 5 I ISSUE 2 I APRIL – JUNE 2018] e ISSN 2348 –1269, Print ISSN 2349-5138 http://ijrar.com/ Cosmos Impact Factor 4.236 ANALYSIS OF WEB LOG DATA USING APACHE PIG IN HADOOP A. C. Priya Ranjani* & Dr. M. Sridhar** *Research Scholar, Department of Computer Science, Acharya Nagarjuna University, Guntur, Andhra Pradesh, INDIA, **Associate Professor, Department of Computer Applications, R.V.R & J.C College of Engineering, Guntur, India Received: April 09, 2018 Accepted: May 22, 2018 ABSTRACT The wide spread use of internet and increased web applications accelerate the rampant growth of web content. Every organization produces huge amount of data in different forms like text, audio, video etc., from multiplesources. The log data stored in web servers is a great source of knowledge. The real challenge for any organization is to understand the behavior of their customers. Analyzing such web log data will help the organizations to understand navigational patterns and interests of their users. As the logs are growing in size day by day, the existing database technologies face a bottleneck to process such massive unstructured data. Hadoop provides a best solution to this problem. Hadoop framework comes up with Hadoop Distributed File System, a reliable distributed storage for data and MapReduce, a distributed parallel processing for executing large volumes of complex data. Hadoop ecosystem constitutes of several other tools like Pig, Hive, Flume, Sqoop etc., for effective analysis of web log data. To write scripts in Map Reduce, one should acquire a good programming knowledge in Java. However Pig, a simple dataflow language can be easily used to analyze such data. -

Apache Pig's Optimizer
Apache Pig’s Optimizer Alan F. Gates, Jianyong Dai, Thejas Nair Hortonworks Abstract Apache Pig allows users to describe dataflows to be executed in Apache Hadoop. The distributed nature of Hadoop, as well as its execution paradigms, provide many execution opportunities as well as impose constraints on the system. Given these opportunities and constraints Pig must make decisions about how to optimize the execution of user scripts. This paper covers some of those optimization choices, focussing one ones that are specific to the Hadoop ecosystem and Pig’s common use cases. It also discusses optimizations that the Pig community has considered adding in the future. 1 Introduction Apache Pig [10] provides an SQL-like dataflow language on top of Apache Hadoop [11] [7]. With Pig, users write dataflow scripts in a language called Pig Latin. Pig then executes these dataflow scripts in Hadoop using MapReduce. Providing users with a scripting language, rather than requiring them to write MapReduce pro- grams in Java, drastically decreases their development time and enables non-Java developers to use Hadoop. Pig also provides operators for most common data processing operations, such as join, sort, and aggregation. It would otherwise require huge amounts of effort for a handcrafted Java MapReduce program to implement these operators. Many different types of data processing are done on Hadoop. Pig does not seek to be a general purpose solution for all of them. Pig focusses on use cases where users have a DAG of transformations to be done on their data, involving some combination of standard relational operations (join, aggregation, etc.) and custom processing which can be included in Pig Latin via User Defined Functions, or UDFs, which can be written in Java or a scripting language.1 Pig also focusses on situations where data may not yet be cleansed and normal- ized. -

HDP 3.1.4 Release Notes Date of Publish: 2019-08-26
Release Notes 3 HDP 3.1.4 Release Notes Date of Publish: 2019-08-26 https://docs.hortonworks.com Release Notes | Contents | ii Contents HDP 3.1.4 Release Notes..........................................................................................4 Component Versions.................................................................................................4 Descriptions of New Features..................................................................................5 Deprecation Notices.................................................................................................. 6 Terminology.......................................................................................................................................................... 6 Removed Components and Product Capabilities.................................................................................................6 Testing Unsupported Features................................................................................ 6 Descriptions of the Latest Technical Preview Features.......................................................................................7 Upgrading to HDP 3.1.4...........................................................................................7 Behavioral Changes.................................................................................................. 7 Apache Patch Information.....................................................................................11 Accumulo........................................................................................................................................................... -

Hot Technologies” Within the O*NET® System
Identification of “Hot Technologies” within the O*NET® System Phil Lewis National Center for O*NET Development Jennifer Norton North Carolina State University Prepared for U.S. Department of Labor Employment and Training Administration Office of Workforce Investment Division of National Programs, Tools, & Technical Assistance Washington, DC April 4, 2016 www.onetcenter.org National Center for O*NET Development, Post Office Box 27625, Raleigh, NC 27611 Table of Contents Background ......................................................................................................................... 2 Hot Technologies Identification Procedure ...................................................................... 3 Mine data to collect the top technology related terms ................................................ 3 Convert the data-mined technology terms into O*NET technologies ......................... 3 Organize the hot technologies within the O*NET Tools & Technology Taxonomy ..... 4 Link the hot technologies to O*NET-SOC occupations .............................................. 4 Determine the display of occupations linked to a hot technology ............................... 4 Summary ............................................................................................................................. 5 Figure 1: O*NET Hot Technology Icon .............................................................................. 6 Appendix A: Hot Technologies Identified During the Initial Implementation ................ 7 National Center -
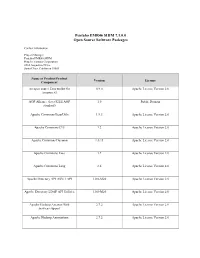
Pentaho EMR46 SHIM 7.1.0.0 Open Source Software Packages
Pentaho EMR46 SHIM 7.1.0.0 Open Source Software Packages Contact Information: Project Manager Pentaho EMR46 SHIM Hitachi Vantara Corporation 2535 Augustine Drive Santa Clara, California 95054 Name of Product/Product Version License Component An open source Java toolkit for 0.9.0 Apache License Version 2.0 Amazon S3 AOP Alliance (Java/J2EE AOP 1.0 Public Domain standard) Apache Commons BeanUtils 1.9.3 Apache License Version 2.0 Apache Commons CLI 1.2 Apache License Version 2.0 Apache Commons Daemon 1.0.13 Apache License Version 2.0 Apache Commons Exec 1.2 Apache License Version 2.0 Apache Commons Lang 2.6 Apache License Version 2.0 Apache Directory API ASN.1 API 1.0.0-M20 Apache License Version 2.0 Apache Directory LDAP API Utilities 1.0.0-M20 Apache License Version 2.0 Apache Hadoop Amazon Web 2.7.2 Apache License Version 2.0 Services support Apache Hadoop Annotations 2.7.2 Apache License Version 2.0 Name of Product/Product Version License Component Apache Hadoop Auth 2.7.2 Apache License Version 2.0 Apache Hadoop Common - 2.7.2 Apache License Version 2.0 org.apache.hadoop:hadoop-common Apache Hadoop HDFS 2.7.2 Apache License Version 2.0 Apache HBase - Client 1.2.0 Apache License Version 2.0 Apache HBase - Common 1.2.0 Apache License Version 2.0 Apache HBase - Hadoop 1.2.0 Apache License Version 2.0 Compatibility Apache HBase - Protocol 1.2.0 Apache License Version 2.0 Apache HBase - Server 1.2.0 Apache License Version 2.0 Apache HBase - Thrift - 1.2.0 Apache License Version 2.0 org.apache.hbase:hbase-thrift Apache HttpComponents Core -

Hadoop Basics.Pdf
Hadoop Illuminated Mark Kerzner <[email protected]> Sujee Maniyam <[email protected]> Hadoop Illuminated by Mark Kerzner and Sujee Maniyam Dedication To the open source community This book on GitHub [https://github.com/hadoop-illuminated/hadoop-book] Companion project on GitHub [https://github.com/hadoop-illuminated/HI-labs] i Acknowledgements From Mark I would like to express gratitude to my editors, co-authors, colleagues, and bosses who shared the thorny path to working clusters - with the hope to make it less thorny for those who follow. Seriously, folks, Hadoop is hard, and Big Data is tough, and there are many related products and skills that you need to master. Therefore, have fun, provide your feedback [http://groups.google.com/group/hadoop-illuminated], and I hope you will find the book entertaining. "The author's opinions do not necessarily coincide with his point of view." - Victor Pelevin, "Generation P" [http://lib.udm.ru/lib/PELEWIN/pokolenie_engl.txt] From Sujee To the kind souls who helped me along the way Copyright © 2013 Hadoop illuminated LLC. All Rights Reserved. ii Table of Contents 1. Who is this book for? ...................................................................................................... 1 1.1. About "Hadoop illuminated" ................................................................................... 1 2. About Authors ................................................................................................................ 2 3. Why do I Need Hadoop ? ................................................................................................