Wordperfect Office Document
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

EGYPTIAN AGRICULTURAL MECHANIZATION PROJECT Contract Number 263-0031-HHC-01
EGYPTIAN AGRICULTURAL MECHANIZATION PROJECT Contract Number 263-0031-HHC-01 ACTIVITY REPORT NUMBER 11 1 October 1983 - 31 December 1983 Submitted by LOUIS BERGER INTERNATIONAL, INC. 100 HalsteadStreet East Orange, New Jersey TABLE OF CONTENTS 1. Summary 1 2. Project Accomplishments 5 3. Financial and Technical Level of Effort 12 4. Implementation 16 5. Next Quarter's Objectives 20 LIST OF FIGURES Figure 1.1 Comparison of budgeted and actual 2 expenditures. Figure 1.2 Credit funds. 2 Figure 4.1 Extension schedule (Jan'84-Jan'85). 17 Figure 4.2 Research schedule (Oct'83-Sept'84). 18 Figure 4.3 Land Improvement schedule 19 (Oct'83-Sept 84). LIST OF TABLES Table 2.0 Demonstration/training equipment: 8 on-site (1983) and planned (1984). Table 2.1 Service center/village workshop loans 10 in-process at Governate banks and at the Project-level, 31/12/83. Table 3.1 Financial level of effort: foreign and 13 local currencies, 15/9/80 - 31/12/84. Table 3.2 Technical level of effort, 14 15/9/80 - 31/12/84. ANNEXES 22 'Annex A Monthly Reports Annex B Machinery Evaluation Series #2: 196 Grain drills, Mower-binders, Combines. Dr. Peter Reiss Annex C Egyptian Research and Development Needs, 225 September, 1983 - December, 1988. Dr. Carl A. Reaves Annex D Computer/Data Acquisition System for the 244 Agricultural Mechanization Research Institute. Dr. M. Yousary Hamdy Annex E A Mechanization Extension Program 273 for the Small Farmer Production Project. Mr. Fred Schantz - 1 1.0 SUMMARY Compared to the budgetary projections for this year (figure 1.1), Project expenditures are on schedules 1) the overall/outgoing category exceeded expectations by 11 per. -

Soil Spatial Variability in Arable Land South of Lake Idku, North-West Nile
id10403671 pdfMachine by Broadgun Software - a great PDF writer! - a great PDF creator! - http://www.pdfmachine.com http://www.broadgun.com EEnnvviirroonnImSmSN : e0e97nn4 - 7tt45aa1 ll SSccVioilueemen n9 Isccsueee 10 An Indian Journal Current Research Paper ESAIJ, 9(10), 2014 [325-344] Soil spatial variability in arable land south of Lake Idku, North- West Nile Delta, Egypt Farida M.S.E.El-Dars1*, Waleed A.Salem2, Mahmoud M.Fahim2 1Department of Chemistry, Faculty of Science, Helwan University, Ain Helwan, Cairo 11795, (EGYPT) 2Soil, Water and Environment Research Institute, Agricultural Research Center, Ministry of Agriculture and Land Reclamation, Giza, (EGYPT) E-mail : [email protected] ABSTRACT Lake Idku (or Idku lagoon) is the third largest coastal water body northwest of the Nile delta located within El Beheira Governorate. Since mid 1950s, over 30% of Lake Idku was dried to create new agricultural lands. The project was extended in 1960s to include areas west of the delta and south of the new/old reclaimed lands. The first study concerning the quality of soil within the study area, south of Lake Idku, was conducted in 1960 and since then, no updates were undertaken to evaluate the effect of the ongoing agricultural/human activities and land use upon the quality of these reclaimed soils. The main objective of this study is to evaluate the change in soil quality in the area relative to the availed analytical data, chemical composition of irrigation water used and crops cultivated during the fall (2010/2011). The impact of different land uses and activities upon the soil quality in the study area, South of Lake Idku, was determined using multivariate analysis (Hierarchal Clustering) in order to identify similarity in patterns and classifying relationships among the measured soil variables. -
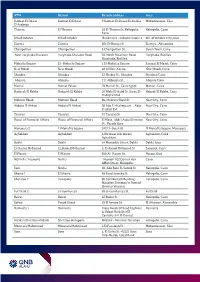
ATM Branch Branch Address Area Gameat El Dowal El
ATM Branch Branch address Area Gameat El Dowal Gameat El Dowal 9 Gameat El-Dewal El-Arabia Mohandessein, Giza El Arabeya Thawra El-Thawra 18 El-Thawra St. Heliopolis, Heliopolis, Cairo Cairo 6th of October 6th of October Banks area - industrial zone 4 6th of October City, Giza Zizenia Zizenia 601 El-Horaya St Zizenya , Alexandria Champollion Champollion 5 Champollion St., Down Town, Cairo New Hurghada Sheraton Hurghada Sheraton Road 36 North Mountain Road, Hurghada, Red Sea Hurghada, Red Sea Mahatta Square El - Mahatta Square 1 El-Mahatta Square Sarayat El Maadi, Cairo New Maadi New Maadi 48 Al Nasr Avenu New Maadi, Cairo Shoubra Shoubra 53 Shobra St., Shoubra Shoubra, Cairo Abassia Abassia 111 Abbassia St., Abassia Cairo Manial Manial Palace 78 Manial St., Cairo Egypt Manial , Cairo Hadayek El Kobba Hadayek El Kobba 16 Waly El-Aahd St, Saray El- Hdayek El Kobba, Cairo Hadayek Mall Makram Ebeid Makram Ebeid 86, Makram Ebeid St Nasr City, Cairo Abbass El Akkad Abbass El Akkad 20 Abo El Ataheya str. , Abas Nasr City, Cairo El akad Ext Tayaran Tayaran 32 Tayaran St. Nasr City, Cairo House of Financial Affairs House of Financial Affairs El Masa, Abdel Azziz Shenawy Nasr City, Cairo St., Parade Area Mansoura 2 El Mohafza Square 242 El- Guish St. El Mohafza Square, Mansoura Aghakhan Aghakhan 12th tower nile towers Aghakhan, Cairo Aghakhan Dokki Dokki 64 Mossadak Street, Dokki Dokki, Giza El- Kamel Mohamed El_Kamel Mohamed 2, El-Kamel Mohamed St. Zamalek, Cairo El Haram El Haram 360 Al- Haram St. Haram, Giza NOZHA ( Triumph) Nozha Triumph.102 Osman Ebn Cairo Affan Street, Heliopolis Safir Nozha 60, Abo Bakr El-Seddik St. -

TO SOME TRACE ELEMENTS UNDER SALT STRESS THROUGHOUT THEIR GROWTH LIFE SPANE: 1- MORPHOLOGICAL and GROWTH CHARACTERS Helaly, M
J. Agric. Sci. Mansoura Univ., 34 (8): 9015 - 9036, 2009 RESPONSE OF TWO NEWLY DEVELOPED CORN GENOTYPE (Zea mays, L) TO SOME TRACE ELEMENTS UNDER SALT STRESS THROUGHOUT THEIR GROWTH LIFE SPANE: 1- MORPHOLOGICAL AND GROWTH CHARACTERS Helaly, M. N. ; A. A. Arafa ; R. A. Fouda and H. F. Esmail Dept. of Agric. Botany, Fac. of Agric., Mansoura Univ. ABSTRACT Two pot experiments were carried out at the greenhouse and laboratories of the Agricultural Botany Department, Faculty of Agriculture, Mansoura University, Egypt during the two seasons of 2002 and 2003 to study to what extent the experimented treatments of some trace-elements applied either as a spray or as a soil feeding may improve salt tolerance and reduce of salt induced necrosis in maize plants; Zea mays, L. Two newly cultivars were chosen; Bachaier 13 single cross and TWC 310 denoted cultivar (1) and cultivar (2), respectively. Four levels of artificial saline sea water having E.C. values of 0.36, 2.5, 5 and 7.5 dsm-1 were used and denoted control, low, high and highest level respectively. Zn, Fe and /or Mn trace-elements were investigated. The experiments were laid out as a factorial complete randomized block design system with 8 replication, four of them were used for the morphological as well as growth characters and analyzed statistically. The general observation was the higher values of growth parameters in cultivar (1) than those obtained with cultivar (2) throughout the experimental period during the two growing seasons. The plants in the two cultivars were growing slowly under salinity and were dwarfing, stunting, inferior in size and unhealthy in general appearance. -

EGYPT Arab Republic of Egypt Ministry of Agriculture and Land Reclamation National Gene Bank and Genetic Resources
COUNTRY REPORT ON THE STATE OF PLANT GENETIC RESOURCES FOR FOOD AND AGRICULTURE ARAB REPUBLIC OF EGYPT Arab Republic of Egypt Ministry of Agriculture and Land Reclamation National Gene Bank and Genetic Resources Second Report on the State of Plant Genetic Resources in the Arab Republic of Egypt March 2007 Note by FAO This Country Report has been prepared by the national authorities in the context of the preparatory process for the Second Report on the State of World’s Plant Genetic Resources for Food and Agriculture. The Report is being made available by the Food and Agriculture Organization of the United Nations (FAO) as requested by the Commission on Genetic Resources for Food and Agriculture. However, the report is solely the responsibility of the national authorities. The information in this report has not been verified by FAO, and the opinions expressed do not necessarily represent the views or policy of FAO. The designations employed and the presentation of material in this information product do not imply the expression of any opinion whatsoever on the part of FAO concerning the legal or development status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. The mention of specific companies or products of manufacturers, whether or not these have been patented, does not imply that these have been endorsed or recommended by FAO in preference to others of a similar nature that are not mentioned. The views expressed in this information product are those of the author(s) and do not necessarily reflect the views of FAO. -

Journal of American Science 2015;11(11)
Journal of American Science 2015;11(11) http://www.jofamericanscience.org Environmental Impact on Water Resources at the Northwestern Part of the Nile Delta, Egypt Awad S.R.1, El Fakharany M.A2 and Hagran N.M.3 1Research Institute for Groundwater, El-Kanater El-Khairia, Egypt 2Department of Geology, Faculty of Science, Benha University, Egypt. 3Drinking Water Supply Company, Giza, Egypt. [email protected] Abstract: Surface water and groundwater in the northwestern part of the Nile Delta is of vital importance for domestic, agricultural, and industrial water supply. The increases in population together with the agricultural and industrial development have resulted in increasing pollution of water resources. The main objective of the present study is to assess the environmental impact on the quality of water resources in the study area. To achieve that, surface water and groundwater samples have been analyzed for major ions, nitrates, and trace elements, in addition to detection of total and fecal coli-form bacteria. The distribution maps for different pollutants in groundwater are carefully studied. Results indicate that both surface water and groundwater in the study area are suffering from quality problems. The majority of the studied area are characterized by fresh water (TDS <1000 ppm) with low contents of Na, Cl, and SO4 and the groundwater is suitable for drinking. High contents are recoded mostly at the northwestern portions as local polluted zones distributed within the area. This is referred to extensive withdrawing in the newly reclaimed areas in the northwestern portions and to the existence of local pollution sources resulted from the infiltration of domestic, agricultural and industrial wastes in the intensive populated area. -

Legislative Analysis to Support Sustainable Approaches to City
People wait for the tram at the station, Alexandria, Egypt @ eFesenko / Shutterstock.com Legislative Analysis to Support Sustainable approaches to City Planning and Extension in Egypt LEGISLATIVE ANALYSIS TO SUPPORT SUSTAINABLE ii APPROACHES TO CITY PLANNING AND EXTENSION IN EGYPT LEGISLATIVE ANALYSIS TO SUPPORT SUSTAINABLE APPROACHES TO CITY PLANNING AND EXTENSION IN EGYPT iii LEGISLATIVE ANALYSIS TO SUPPORT SUSTAINABLE APPROACHES TO CITY PLANNING AND EXTENSION IN EGYPT LEGISLATIVE ANALYSIS TO SUPPORT SUSTAINABLE iv APPROACHES TO CITY PLANNING AND EXTENSION IN EGYPT Copyright © United Nations Human Settlements Programme (UN-Habitat), 2015 December, 2015 Disclaimer The designations employed and the presentation of the material in this document do not imply the expression of any opinion whatsoever on the part of the Secretariat off the United Nations concerning the legal status of any country, territory, city or area, or of its authorities, or concerning delimitation of its frontiers or boundaries, or regarding its economic system or degree of development. The analysis, conclusions and recommendations of this document do not necessarily reflect the views of the United Nations Human Settlements Programme, the Governing Council of the United Nations Human Settlements Programme or its Member States. References to names of firms and commercial products and processes does not imply their endorsement by the United Nations, and a failure to mention a particular firm, commercial product or process is not a sign of disapproval. Excerpts from the -

The-Second-Republic E 2021-1.Pdf
Dr. Khaled Okasha General Manager Dr. Abdel-Moneim Said Academic advisor Editor Belal Manzour Translators Asmaa El-Shafei Omnia El-Sayed Mariam Khaled General Coordinator Mai Saeed Art direction Eslam Ali Index Introduction: 4 Building the Egyptian Human Being I. Developing the Health Sector 6 II. Developing the Education Sector 12 III. Economic Policies 16 IV. Political Life 22 V. Developing Border Regions 24 VI. Fourth-Generation Cities 28 VII. Developing the Transport System 32 VIII. Developments in the Security and 36 Military Field IX. Egypt’s Foreign Policy 40 X. Energy Projects and the East 46 Mediterranean Gas Forum XI. Egypt’s Role in Africa 52 Research Team (Alphabetically) Ahmed Bayoumi Heba Zean Dr. Mohamed Shadi Amal Ismail Hussein Abd Elrady Mohammed Mansour Asmaa Fahmy Kenzy Serge Moustafa Abdella Basant Gamal Mahmoud Qassem Dr. Omar El-Hosseiny Ferdous Abdelbaki Mohamed Abdelrazeq Rahma Hassan Hala Fouda Mohamed Hassan Tokka ELnaggar Introduction Building the Egyptian Human Being gypt has been taking solid resolute steps toward drafting a new social contract since the 30 June 2013 Revolution. The contract addresses the mistakes of the past, all the Ewhile working according to a strategy that takes into account several security, polit- ical, economic and social factors. In this context, the concept of the New Republic represents a comprehensive framework and the philosophy adopted by the Egyptian state, based on a set of principles that are founded on two main pillars. The first is that socioeconomic development is a prerequisite for political development, which is the foundation for the stability of the state and its institutions and the sustainability of the social fabric. -

Accessibility Impact Analysis of New Public Transit Projects in Cairo, Egypt
Accessibility Impact Analysis of New Public Transit Projects in Cairo, Egypt By Adham Kalila B.Eng in Civil Engineering McGill University Montreal, Canada (2012) Master of Science in Transportation Massachusetts Institute of Technology Cambridge, Massachusetts (2018) Submitted to the Department of Urban Studies and Planning in partial fulfillment of the requirements for the degree of Master of Science in Urban Planning at the MASSACHUSETTS INSTITUTE OF TECHNOLOGY June 2019 © 2019 Adham Kalila. All Rights Reserved The author hereby grants to MIT the permission to reproduce and to distribute publicly paper and electronic copies of the thesis document in whole or in part in any medium now known or hereafter created. Author________________________________________________________________________ Department of Urban Studies and Planning 05/21/2019 Certified by____________________________________________________________________ Professor Sarah Williams Department of Urban Studies and Planning Thesis Supervisor Accepted by___________________________________________________________________ Professor of the Practice, Caesar McDowell Co-Chair, MCP Committee Department of Urban Studies and Planning Accessibility Impact Analysis of New Public Transit Projects in Cairo, Egypt By Adham Kalila Submitted to the Department of Urban Studies and Planning on May 21, 2019 in Partial Fulfillment of the Requirements for the Degree of Master of Science in Urban Planning Abstract The New Urban Communities (NUC), built around Cairo, developed to relieve congestion -

Egyptian Electricity Holding Company
Egyptian Electricity Holding Company 2 Annual Report 2015 / 2016 Contents Vision & Mission ............................................................................................................................................................ 5 Egyptian Electricity Holding Company .......................................................................................................... 7 Organizational Structure of EEHC ................................................................................................................... 8 Board of Directors of EEHC .................................................................................................................................. 9 Introduction ...................................................................................................................................................................... 10 Electricity for 2015/2016 ........................................................................................................................................... 13 Financial Position of EEHC and its affiliated Companies ................................................ 15 Electric Power Production ....................................................................................................................................... 17 - Installed Capacities of Power Stations Statistics (30/6/2016) ................................................... 18 - Generated and Purchased Energy ........................................................................................................... -

Arab Republic of Egypt Towards an Urban
41178 v 2 ARAB REPUBLIC OF EGYPT TOWARDS AN URBAN SECTOR STRATEGY Public Disclosure Authorized Public Disclosure Authorized Public Disclosure Authorized Public Disclosure Authorized THE WORLD BANK June 2008 Report No.: 44506-EG Arab Republic of Egypt Urban Sector Note VOLUME TWO TOWARDS AN URBAN SECTOR STRATEGY JUNE, 2008 Sustainable Development Department Middle East and North Africa Region i Document of the World Bank ii CURRENCY EQUIVALENTS (Exchange Rate Effective June, 2008) Currency Unit = Egyptian Pound (LE) Egyptian Pounds 5.33 = US$1 ABBREVIATIONS AND ACRONYMS BOT Build, Operate, and Transfer CAPMAS Central Agency for Public Mobilization and Statistics CDA Community Development Association CDS City Development Strategy EEAA Egyptian Environmental Affairs Agency ERA Egyptian Railways Authority ESA Egyptian Survey Authority GDP Gross Domestic Product GOE Government of Egypt GOPP General Organization for Physical Planning GTZ German Technical Cooperation LE Livres Egyptiens MENA Middle East and North Africa MFA Mortgage Finance Authority MHUUD Ministry of Housing, Utilities, and Urban Development MOA Ministry of Agriculture MOF Ministry of Finance MOI Ministry of Investment MOLD Ministry of Local Development NCUPD National Council for Urban Planning and Development NHP National Housing Program NGO Non-governmental Organization NUCA New Urban Communities Authority USAID United States Agency for International Development Ziman The official boundary between agricultural and desert land Vice President: Daniela Gressani Country Manager/Director: -

The Project for the Establishment of National and Local Monitoring Systems to Observe and Evaluate Land Development Policies in Egypt
THE PROJECT FOR THE ESTABLISHMENT OF NATIONAL AND LOCAL MONITORING SYSTEMS TO OBSERVE AND EVALUATE LAND DEVELOPMENT POLICIES IN EGYPT - Final Report - 2007. 05 KOICA <PREFACE> In Egypt, 96% of the entire territory is desert, and most of the population is concentrated in the remaining 4%. However, rapid increase in population and urban concentration have caused imbalance between demand and supply of land development. Consequently, unplanned development has been spreading rapidly in the form of informal settlements through illegal land development and farmland conversion. Recognizing this problem, the Egyptian government asked Korea, which has an experience of successfully solving similar land problems in a short period in the recent past, for assistance regarding land policy, urban planning, and policy evaluation and observation capacity. Accordingly, under the control of the Korea International Cooperation Agency, the Korea Research Institute for Human Settlements launched a joint research project with the General Organization for Physical Planning (GOPP) of Egypt. The original project period was 18 months from July 2005 to January 2007 while it took a total of 20 months until March 2007; it was extended by two months with the request from the Egyptian counterpart. Throughout the research project, we endeavored to actively meet the requirements of the Egyptian government from the stage of prior investigation. The official title of the project is 'THE ESTABLISHMENT OF NATIONAL AND LOCAL MONITORING SYSTEMS TO OBSERVE AND EVALUATE LAND DEVELOPMENT POLICIES IN EGYPT'. Various methods were utilized for the research including literature review, onsite investigation of the project areas in Egypt, surveys, case studies of Korea and other countries, education for the GOPP officials in Korea, and meetings and workshops in Egypt.