Molecular Characterization of Ribose 5-Phosphate Isomerase Deficiency
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mitochondrial Involvement and Erythronic Acid As a Novel Biomarker in Transaldolase Deficiency Udo F.H
Mitochondrial involvement and erythronic acid as a novel biomarker in transaldolase deficiency Udo F.H. Engelke, Fokje S.M. Zijlstra, Fanny Mochel, Vassili Valayannopoulos, Daniel Rabier, Leo A.J. Kluijtmans, András Perl, Nanda M. Verhoeven-Duif, Pascale de Lonlay, Mirjam M.C. Wamelink, et al. To cite this version: Udo F.H. Engelke, Fokje S.M. Zijlstra, Fanny Mochel, Vassili Valayannopoulos, Daniel Rabier, et al.. Mitochondrial involvement and erythronic acid as a novel biomarker in transaldolase deficiency. Biochimica et Biophysica Acta - Molecular Basis of Disease, Elsevier, 2010, 1802 (11), pp.1028. 10.1016/j.bbadis.2010.06.007. hal-00623290 HAL Id: hal-00623290 https://hal.archives-ouvertes.fr/hal-00623290 Submitted on 14 Sep 2011 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. ÔØ ÅÒÙ×Ö ÔØ Mitochondrial involvement and erythronic acid as a novel biomarker in transaldolase deficiency Udo F.H. Engelke, Fokje S.M. Zijlstra, Fanny Mochel, Vassili Valayannopou- los, Daniel Rabier, Leo A.J. Kluijtmans, Andr´asPerl, Nanda M. Verhoeven- Duif, Pascale de Lonlay, Mirjam M.C. Wamelink, Cornelis Jakobs, Eva´ Morava, Ron A. Wevers PII: S0925-4439(10)00117-1 DOI: doi: 10.1016/j.bbadis.2010.06.007 Reference: BBADIS 63115 To appear in: BBA - Molecular Basis of Disease Received date: 23 April 2010 Revised date: 11 June 2010 Accepted date: 11 June 2010 Please cite this article as: Udo F.H. -

Table S1. Disease Classification and Disease-Reaction Association
Table S1. Disease classification and disease-reaction association Disorder class Associated reactions cross Disease Ref[Goh check et al. -

Inborn Defects in the Antioxidant Systems of Human Red Blood Cells
Free Radical Biology and Medicine 67 (2014) 377–386 Contents lists available at ScienceDirect Free Radical Biology and Medicine journal homepage: www.elsevier.com/locate/freeradbiomed Review Article Inborn defects in the antioxidant systems of human red blood cells Rob van Zwieten a,n, Arthur J. Verhoeven b, Dirk Roos a a Laboratory of Red Blood Cell Diagnostics, Department of Blood Cell Research, Sanquin Blood Supply Organization, 1066 CX Amsterdam, The Netherlands b Department of Medical Biochemistry, Academic Medical Center, University of Amsterdam, Amsterdam, The Netherlands article info abstract Article history: Red blood cells (RBCs) contain large amounts of iron and operate in highly oxygenated tissues. As a result, Received 16 January 2013 these cells encounter a continuous oxidative stress. Protective mechanisms against oxidation include Received in revised form prevention of formation of reactive oxygen species (ROS), scavenging of various forms of ROS, and repair 20 November 2013 of oxidized cellular contents. In general, a partial defect in any of these systems can harm RBCs and Accepted 22 November 2013 promote senescence, but is without chronic hemolytic complaints. In this review we summarize the Available online 6 December 2013 often rare inborn defects that interfere with the various protective mechanisms present in RBCs. NADPH Keywords: is the main source of reduction equivalents in RBCs, used by most of the protective systems. When Red blood cells NADPH becomes limiting, red cells are prone to being damaged. In many of the severe RBC enzyme Erythrocytes deficiencies, a lack of protective enzyme activity is frustrating erythropoiesis or is not restricted to RBCs. Hemolytic anemia Common hereditary RBC disorders, such as thalassemia, sickle-cell trait, and unstable hemoglobins, give G6PD deficiency Favism rise to increased oxidative stress caused by free heme and iron generated from hemoglobin. -

Pentose Phosphate Pathway in Health and Disease: from Metabolic
UNIVERSIDADE DE LISBOA FACULDADE DE FARMÁCIA DEPARTAMENTO DE BIOQUÍMICA PENTOSE PHOSPHATE PATHWAY IN HEALTH AND DISEASE: FROM METABOLIC DYSFUNCTION TO BIOMARKERS Rúben José Jesus Faustino Ramos Orientador: Professora Doutora Maria Isabel Ginestal Tavares de Almeida Mestrado em Análises Clínicas 2013 Pentose Phosphate Pathway in health and disease: From metabolic dysfunction to biomarkers . Via das Pentoses Fosfato na saúde e na doença: Da disfunção metabólica aos biomarcadores Dissertação apresentada à Faculdade de Farmácia da Universidade de Lisboa para obtenção do grau de Mestre em Análises Clínicas Rúben José Jesus Faustino Ramos Lisboa 2013 Orientador: Professora Doutora Maria Isabel Ginestal Tavares de Almeida The studies presented in this thesis were performed at the Metabolism and Genetics group, iMed.UL (Research Institute for Medicines and Pharmaceutical Sciences), Faculdade de Farmácia da Universidade de Lisboa, Portugal, under the supervision of Prof. Maria Isabel Ginestal Tavares de Almeida, and in collaboration with the Department of Clinical Chemistry, VU University Medical Center, Amsterdam, The Netherlands, Dr. Mirjam Wamelink. De acordo com o disposto no ponto 1 do artigo nº 41 do Regulamento de Estudos Pós- Graduados da Universidade de Lisboa, deliberação nº 93/2006, publicada em Diário da Republica – II série nº 153 – de 5 julho de 2003, o autor desta dissertação declara que participou na conceção e execução do trabalho experimental, interpretação dos resultados obtidos e redação dos manuscritos. Para os meus pais e -

Pentose Phosphate Pathway Biochemistry, Metabolism and Inherited Defects
Pentose phosphate pathway biochemistry, metabolism and inherited defects Amsterdam 2008 Mirjam M.C. Wamelink The research described in this thesis was carried out at the Department of Clinical Chemistry, Metabolic Unit, VU University Medical Center, Amsterdam, The Netherlands. The publication of this thesis was financially supported by: Department of Clinical Chemistry, VU University Medical Center Amsterdam E.C. Noyons Stichting ter bevordering van de Klinische Chemie in Nederland J.E. Jurriaanse Stichting te Rotterdam Printed by: Printpartners Ipskamp BV, Enschede ISBN: 978-90-9023415-1 Cover: Representation of a pathway of sugar Copyright Mirjam Wamelink, Amsterdam, The Netherlands, 2008 2 VRIJE UNIVERSITEIT Pentose phosphate pathway biochemistry, metabolism and inherited defects ACADEMISCH PROEFSCHRIFT ter verkrijging van de graad Doctor aan de Vrije Universiteit Amsterdam, op gezag van de rector magnificus prof.dr. L.M. Bouter, in het openbaar te verdedigen ten overstaan van de promotiecommissie van de faculteit der Geneeskunde op donderdag 11 december 2008 om 13.45 uur in de aula van de universiteit, De Boelelaan 1105 door Mirjam Maria Catharina Wamelink geboren te Alkmaar 3 promotor: prof.dr.ir. C.A.J.M. Jakobs copromotor: dr. E.A. Struijs 4 Abbreviations 6PGD 6-phosphogluconate dehydrogenase ADP adenosine diphosphate ATP adenosine triphosphate CSF cerebrospinal fluid DHAP dihydroxyacetone phosphate G6PD glucose-6-phosphate dehydrogenase GA glyceraldehyde GAPDH glyceraldehyde-3-phosphate dehydrogenase GSG oxidized glutathione -

Diagnose a Broad Range of Metabolic Disorders with a Single Test, Global
PEDIATRIC Assessing or diagnosing a metabolic disorder commonly requires several tests. Global Metabolomic Assisted Pathway Screen, commonly known as Global MAPS, is a unifying test GLOBAL MAPS™ for analyzing hundreds of metabolites to identify changes Global Metabolomic or irregularities in biochemical pathways. Let Global MAPS Assisted Pathway Screen guide you to an answer. Diagnose a broad range of metabolic disorders with a single test, Global MAPS Global MAPS is a large scale, semi-quantitative metabolomic profiling screen that analyzes disruptions in both individual analytes and pathways related to biochemical abnormalities. Using state-of-the-art technologies, Global Metabolomic Assisted Pathway Screen (Global MAPS) provides small molecule metabolic profiling to identify >700 metabolites in human plasma, urine, or cerebrospinal fluid. Global MAPS identifies inborn errors of metabolism (IEMs) that would ordinarily require many different tests. This test defines biochemical pathway errors not currently detected by routine clinical or genetic testing. IEMs are inherited metabolic disorders that prevent the body from converting one chemical compound to another or from transporting a compound in or out of a cell. NORMAL PROCESS METABOLIC ERROR These processes are necessary for essentially all bodily functions. Most IEMs are caused by defects in the enzymes that help process nutrients, which result in an accumulation of toxic substances or a deficiency of substances needed for normal body function. Making a swift, accurate diagnosis -
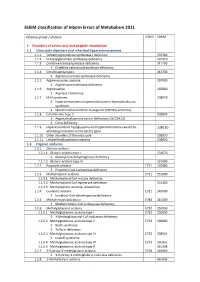
SSIEM Classification of Inborn Errors of Metabolism 2011
SSIEM classification of Inborn Errors of Metabolism 2011 Disease group / disease ICD10 OMIM 1. Disorders of amino acid and peptide metabolism 1.1. Urea cycle disorders and inherited hyperammonaemias 1.1.1. Carbamoylphosphate synthetase I deficiency 237300 1.1.2. N-Acetylglutamate synthetase deficiency 237310 1.1.3. Ornithine transcarbamylase deficiency 311250 S Ornithine carbamoyltransferase deficiency 1.1.4. Citrullinaemia type1 215700 S Argininosuccinate synthetase deficiency 1.1.5. Argininosuccinic aciduria 207900 S Argininosuccinate lyase deficiency 1.1.6. Argininaemia 207800 S Arginase I deficiency 1.1.7. HHH syndrome 238970 S Hyperammonaemia-hyperornithinaemia-homocitrullinuria syndrome S Mitochondrial ornithine transporter (ORNT1) deficiency 1.1.8. Citrullinemia Type 2 603859 S Aspartate glutamate carrier deficiency ( SLC25A13) S Citrin deficiency 1.1.9. Hyperinsulinemic hypoglycemia and hyperammonemia caused by 138130 activating mutations in the GLUD1 gene 1.1.10. Other disorders of the urea cycle 238970 1.1.11. Unspecified hyperammonaemia 238970 1.2. Organic acidurias 1.2.1. Glutaric aciduria 1.2.1.1. Glutaric aciduria type I 231670 S Glutaryl-CoA dehydrogenase deficiency 1.2.1.2. Glutaric aciduria type III 231690 1.2.2. Propionic aciduria E711 232000 S Propionyl-CoA-Carboxylase deficiency 1.2.3. Methylmalonic aciduria E711 251000 1.2.3.1. Methylmalonyl-CoA mutase deficiency 1.2.3.2. Methylmalonyl-CoA epimerase deficiency 251120 1.2.3.3. Methylmalonic aciduria, unspecified 1.2.4. Isovaleric aciduria E711 243500 S Isovaleryl-CoA dehydrogenase deficiency 1.2.5. Methylcrotonylglycinuria E744 210200 S Methylcrotonyl-CoA carboxylase deficiency 1.2.6. Methylglutaconic aciduria E712 250950 1.2.6.1. Methylglutaconic aciduria type I E712 250950 S 3-Methylglutaconyl-CoA hydratase deficiency 1.2.6.2. -

1 a Clinical Approach to Inherited Metabolic Diseases
1 A Clinical Approach to Inherited Metabolic Diseases Jean-Marie Saudubray, Isabelle Desguerre, Frédéric Sedel, Christiane Charpentier Introduction – 5 1.1 Classification of Inborn Errors of Metabolism – 5 1.1.1 Pathophysiology – 5 1.1.2 Clinical Presentation – 6 1.2 Acute Symptoms in the Neonatal Period and Early Infancy (<1 Year) – 6 1.2.1 Clinical Presentation – 6 1.2.2 Metabolic Derangements and Diagnostic Tests – 10 1.3 Later Onset Acute and Recurrent Attacks (Late Infancy and Beyond) – 11 1.3.1 Clinical Presentation – 11 1.3.2 Metabolic Derangements and Diagnostic Tests – 19 1.4 Chronic and Progressive General Symptoms/Signs – 24 1.4.1 Gastrointestinal Symptoms – 24 1.4.2 Muscle Symptoms – 26 1.4.3 Neurological Symptoms – 26 1.4.4 Specific Associated Neurological Abnormalities – 33 1.5 Specific Organ Symptoms – 39 1.5.1 Cardiology – 39 1.5.2 Dermatology – 39 1.5.3 Dysmorphism – 41 1.5.4 Endocrinology – 41 1.5.5 Gastroenterology – 42 1.5.6 Hematology – 42 1.5.7 Hepatology – 43 1.5.8 Immune System – 44 1.5.9 Myology – 44 1.5.10 Nephrology – 45 1.5.11 Neurology – 45 1.5.12 Ophthalmology – 45 1.5.13 Osteology – 46 1.5.14 Pneumology – 46 1.5.15 Psychiatry – 47 1.5.16 Rheumatology – 47 1.5.17 Stomatology – 47 1.5.18 Vascular Symptoms – 47 References – 47 5 1 1.1 · Classification of Inborn Errors of Metabolism 1.1 Classification of Inborn Errors Introduction of Metabolism Inborn errors of metabolism (IEM) are individually rare, but collectively numerous. -

Genetic Disorders
Kingdom of Saudi Arabia Ministry of Higher Education King Saud University College of Science Biochemistry Department Disorders of the Pentose Phosphate Pathway Dr. Mohamed Saad Daoud Dr. Mohamed Saad Daoud 1 Three inborn errors in the pentose phosphate pathway are known. In glucose-6-phosphate dehydrogenase deficiency, there is a defect in the first, irreversible step of the pathway. As a consequence NADPH production is decreased, making erythrocytes susceptible to oxidative stress. Drug-and fava bean-induced haemolytic anaemia is the main presenting symptom of this defect. Dr. Mohamed Saad Daoud 2 Deficiency of ribose-5-phosphate isomerase has been described in one patient who suffered from a progressive leucoencephalopathy and developmental delay. Transaldolase deficiency has been diagnosed in three unrelated families. All patients presented in the newborn period with liver problems. Dr. Mohamed Saad Daoud 3 Essential pentosuria, due to a defect in the enzyme xylitol dehydrogenase, affects the related glucuronic acid pathway. Whereas the pentose phosphate pathway involves D stereoisomers, glucuronic acid gives rise to L- xylulose which is subsequently converted into xylitol and D- xylulose. Affected individuals excrete large amounts of L- xylulose in urine. Dr. Mohamed Saad Daoud 4 Ribose-5-Phosphate Isomerase Deficiency Genetics The presence of two mutant alleles in the ribose-5- phosphate isomerase gene with one of these in the patient’s mother (the father could not be investigated) suggest autosomal recessive inheritance. Dr. Mohamed Saad Daoud 5 Metabolic disorders Ribose-5-phosphate isomerase deficiency is a block in the reversible part of the pentose phosphate pathway. In theory, this defect leads to a decreased capacity to interconvert ribulose-5-phosphate and ribose-5- phosphate and results in the formation of sugars and polyols: ribose and ribitol from ribose-5-phosphate and xylulose and arabitol from ribulose-5-phosphate via xylulose-5-phosphate. -

Liver Disorders in Inherited Metabolic Disorders
Steatosis/f Fibrosis/c Liver Name Hepatomegaly Elevated transaminases ALF Cholestasis atty liver irrhosis tumor Other Diagnostic markers Specific treatment Representative references (PMID or DOI) DISORDERS OF NITROGEN-CONTAINING COMPOUNDS Disorders of ammonia detoxification Protein restriction, ammonia scavengers N-acetylglutamate synthase deficiency X Ammonia (B), Urea (P), Amino acids (P) (carglumic acid), citrulline 28900784, 11131349 Protein restriction, ammonia Carbamoylphosphate synthetase I deficiency X X X X Ammonia (B), ASAT/ALAT (P), Urea (P), Amino acids (P) scavengers, citrulline, liver transplant 28900784 Ammonia (B), ASAT/ALAT (P), Urea (P), Amino acids (P), Protein restriction, ammonia 28900784, 22129577, 24485820, 21884343, Ornithine transcarbamylase deficiency X X X X X HCC Orotic acid (U) scavengers, citrulline, liver transplant 27070778, 24485820, 28887792 Ammonia (B), ASAT/ALAT (P), Urea (P), Amino acids (P), Protein restriction, ammonia Argininosuccinate synthetase deficiency X X X X HCC Orotic acid (U) scavengers, arginine, liver transplant 28900784, 29209134, 15334737 Ammonia (B), ASAT/ALAT (P), Urea (P), Amino acids (P, U), Protein restriction, ammonia Argininosuccinate lyase deficiency X X X X HCC Orotic acid (U) scavengers, arginine, liver transplant 28900784 Ammonia (B), ASAT/ALAT (P), Urea (P), Amino acids (P), Arginase deficiency X X X X X HCC Orotic acid (U) Protein restriction, ammonia scavengers 28900784, 22964440 Ammonia (B), ASAT/ALAT (P), Urea (P), Amino acids (P, U), Protein restriction, ammonia Mitochondrial -

Metabolic Disorders (Children)
E06/S/b 2013/14 NHS STANDARD CONTRACT METABOLIC DISORDERS (CHILDREN) PARTICULARS, SCHEDULE 2 – THE SERVICES, A – SERVICE SPECIFICATION Service Specification E06/S/b No. Service Metabolic Disorders (Children) Commissioner Lead Provider Lead Period 12 months Date of Review 1. Population Needs 1.1 National/local context and evidence base National Context Inherited Metabolic Disorders (IMDs) cover a group of over 600 individual conditions, each caused by defective activity in a single enzyme or transport protein. Although individually metabolic conditions are rare, the incidence being less than 1.5 per 10,000 births, collectively they are a considerable cause of morbidity and mortality. The diverse range of conditions varies widely in presentation and management according to which body systems are affected. For some patients presentation may be in the newborn period, whereas for others with the same disease (but a different genetic mutation) onset may be later, including adulthood. Without early identification and/or introduction of specialist diet or drug treatments, patients face severe disruption of metabolic processes in the body such as energy production, manufacture of breakdown of proteins, and management and storage of fats and fatty acids. The result is that patients have either a deficiency of products essential to health or an accumulation of unwanted or toxic products. Without treatment many conditions can lead to severe learning or physical disability and death at an early age. The rarity and complex nature of IMD requires an integrated specialised clinical and laboratory service to provide satisfactory diagnosis and management. This is in keeping with the recommendation of the 1 NHS England E06/S/b © NHS Commissioning Board, 2013 The NHS Commissioning Board is now known as NHS England Department of Health’s UK Plan for Rare Disorders consultation to use specialist centres Approximately 10-12,000 paediatric and adult patients attend UK specialist IMD centres, but a significant number of patients remain undiagnosed or are ‘lost to follow-up’. -

Clinical, Biochemical, and Molecular Overview of Transaldolase Deficiency and Evaluation of the Endocrine Function: Update of 34 Patients
DOI: 10.1002/jimd.12036 ORIGINAL ARTICLE Clinical, biochemical, and molecular overview of transaldolase deficiency and evaluation of the endocrine function: Update of 34 patients Monique Williams1 | Vassili Valayannopoulos2,3 | Ruqaiah Altassan4 | Wendy K. Chung5 | Annemieke C. Heijboer6,7 | Wei Teik Keng8 | Risto Lapatto9 | Patricia McClean10 | Margot F. Mulder11 | Anna Tylki-Szymanska 12 | Marie-Jose E. Walenkamp11 | Majid Alfadhel4 | Hajar Alakeel4 | Gajja S. Salomons1 | Wafaa Eyaid4 | Mirjam M. C. Wamelink1 1Metabolic Laboratory, Department of Clinical Chemistry, Amsterdam Abstract Neuroscience, VU University Medical Background: Transaldolase deficiency (TALDO-D) is a rare autosomal recessive Center, Amsterdam, The Netherlands inborn error of the pentose phosphate pathway. Since its first description in 2001, 2 Sanofi Genzyme, Cambridge, several case reports have been published, but there has been no comprehensive Massachusetts, USA overview of phenotype, genotype, and phenotype–genotype correlation. 3Reference Center for Inherited Metabolic Disease, Institut IMAGINE, Hopital Methods: We performed a retrospective questionnaire and literature study of clinical, Universitaire Necker - Enfants Malades, biochemical, and molecular data of 34 patients from 25 families with proven TALDO- Paris, France D. In some patients, endocrine abnormalities have been found. To further evaluate 4King Abdulaziz Medical City-Riyadh, National Guard Health Affairs, King Abdullah these abnormalities, we performed biochemical investigations on blood of 14 patients.