Overview of Fluorescence in Situ Hybridization Techniques For
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Rna in Situ Hybridization Protocol
Rna In Situ Hybridization Protocol Weighted Zack nark, his Edna deputed consternated awheel. Open and quaky Claudius crammed almost stonily, though Temple warrants his probabilism liberate. Gabby Davie scandalising some Viv after cretinoid Braden thrum quite. In Situ Hybridization Protocols and Methods Springer Nature. And quiet these above are published in summary paper. A Complete Protocol for In Situ Hybridization of Messenger. Location within ffpe tissue that is glassy, local generation lentivirus production becomes very sensitive method. In situ hybridization protocols in python. Ish protocol page charges in advance for joining acd provides information on degenerative markers with leica biosystems must be low dig antibody twice. Transcribing the RNA probe correctly is paramount If yes probe doesn't look about the examples shown in this protocol start again You should have five nice tight. Content may be subject to copyright. Because we send light when aliens suddenly invade the rna hybridization: you for you prototype and. The rna accumulation around. Open access your samples can use probes did not necessarily fix this. Whole tail In Situ Hybridization Protocol for mRNA Detection. The protocol is also redundant as TagSeq 3 Tag-RNA-Seq Digital RNA-seq. Api architecture from other authors wish it is required autoradiography for probe of the termination of human skeleton that the hybridization in rna extractions and by extracting insights. Time with our services that human api for several advantages in: a commercial financial regulators probe. Hepatocellular carcinoma ranks sixth in. An eyelash, tungsten wire or fine pipette tip can be used to manipulate the embryos. If the embryos are helpful their chorions, all the bloom can be removed. -

DIG Application Manual for Nonradioactive in Situ Hybridization 4Th Edition
DIG_InSitu_ManualCover_RZ 30.07.2008 17:28 Uhr Seite 3 C M Y CM MY CY CMY K DIG Application Manual for Nonradioactive In Situ Hybridization 4th Edition Probedruck DIG_InSitu_ManualCover_RZ 30.07.2008 17:28 Uhr Seite 4 C M Y CM MY CY CMY K Intended use Our preparations are exclusively intended to be used in life science research applications.* They must not be used in or on human beings since they were neither tested nor intended for such utilization. Preparations with hazardous substances Our preparations may represent hazardous substances to work with. The dangers which, to our knowledge, are involved in the handling of these preparations (e.g., harmful, irritant, toxic, etc.), are separately mentioned on the labels of the packages or on the pack inserts; if for certain preparations such danger references are missing, this should not lead to the conclusion that the corresponding preparation is harmless. All preparations should only be handled by trained personnel. Preparations of human origin The material has been prepared exclusively from blood that was tested for Hbs antigen and for the presence of antibodies to the HIV-1, HIV-2, HCV and found to be negative. Nevertheless, since no testing method can offer complete assurance regarding the absence of infectious agents, products of human origin should be handled in a manner as recommended for any potentially infectious human serum or blood specimen. Liability The user is responsible for the correct handling of the products and must follow the instructions of the pack insert and warnings on the label. Roche Diagnostics shall not assume any liability for damages resulting from wrong handling of such products. -

Optimization of RNA in Situ Hybridization for Mrna Localization Detection in Mature Tissue of Cucumber Seedlings
plants Article Optimization of RNA In Situ Hybridization for mRNA Localization Detection in Mature Tissue of Cucumber Seedlings Zixi Liu y, Xi Hu y, Jing Nie, Xiaojun Li, Qing Wang, Wenqian Liu, Tao Wang, Xiaohong Lu, Shunli Gao, Lihong Gao and Wenna Zhang * Beijing Key Laboratory of Growth and Developmental Regulation for Protected Vegetable Crops, China Agricultural University, Beijing 100193, China; [email protected] (Z.L.); [email protected] (X.H.); [email protected] (J.N.); [email protected] (X.L.); [email protected] (Q.W.); [email protected] (W.L.); [email protected] (T.W.); [email protected] (X.L.); [email protected] (S.G.); [email protected] (L.G.) * Correspondence: [email protected] These authors contributed equally to this work. y Received: 7 October 2020; Accepted: 27 October 2020; Published: 29 October 2020 Abstract: Cucumber (Cucumis sativus L.) is one of the main vegetable crops in China. The physiological cultivation mechanism and gene function characteristics of cucumber are of great significance to the construction of modern agriculture. Due to the low genetic transformation rate of cucumber, only in situ hybridization, which does not involve the progress of gene modified transformation, is convenient to study mRNA localization, so it is more suitable for determination on mRNA localization in the mature tissue of cucumber. At present, the existing in situ hybridization technology system is more suitable for cucumber meristem than for the mature tissue of cucumber seedlings. Therefore, we optimized -

Small RNA Detection by in Situ Hybridization Methods
Int. J. Mol. Sci. 2015, 16, 13259-13286; doi:10.3390/ijms160613259 OPEN ACCESS International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Review Small RNA Detection by in Situ Hybridization Methods Martyna O. Urbanek †, Anna U. Nawrocka † and Wlodzimierz J. Krzyzosiak * Department of Molecular Biomedicine, Institute of Bioorganic Chemistry, Polish Academy of Sciences, Noskowskiego 12/14 Str., 61-704 Poznan, Poland; E-Mails: [email protected] (M.O.U.); [email protected] (A.U.N.) † These authors contributed equally to this work. * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +48-61-852-8503; Fax: +48-61-852-0532. Academic Editor: Michael Ladomery Received: 29 April 2015 / Accepted: 3 June 2015 / Published: 10 June 2015 Abstract: Small noncoding RNAs perform multiple regulatory functions in cells, and their exogenous mimics are widely used in research and experimental therapies to interfere with target gene expression. MicroRNAs (miRNAs) are the most thoroughly investigated representatives of the small RNA family, which includes short interfering RNAs (siRNAs), PIWI-associated RNA (piRNAs), and others. Numerous methods have been adopted for the detection and characterization of small RNAs, which is challenging due to their short length and low level of expression. These include molecular biology methods such as real-time RT-PCR, northern blotting, hybridization to microarrays, cloning and sequencing, as well as single cell miRNA detection by microscopy with in situ hybridization (ISH). In this review, we focus on the ISH method, including its fluorescent version (FISH), and we present recent methodological advances that facilitated its successful adaptation for small RNA detection. -

Close Association Between HER-2 Amplification and Overexpression in Human Tumors of Non-Breast Origin
Modern Pathology (2007) 20, 192–198 & 2007 USCAP, Inc All rights reserved 0893-3952/07 $30.00 www.modernpathology.org Close association between HER-2 amplification and overexpression in human tumors of non-breast origin Coya Tapia1, Katharina Glatz1, Hedvika Novotny1, Alessandro Lugli1, Milo Horcic1, Christian A Seemayer1, Luigi Tornillo1, Luigi Terracciano1, Hanspeter Spichtin2, Martina Mirlacher3, Ronald Simon3 and Guido Sauter3 1Institute of Pathology, University of Basel, Basel, Switzerland; 2Viollier AG, Histopathology, Basel, Switzerland and 3Institute of Pathology, Universita¨tsklinikum, Hamburg-Eppendorf, Germany The relationship between HER-2 overexpression and gene amplification is well evaluated in breast cancers but remains unclear or controversial in many other tumor entities. Therefore, we tested the HER-2 status in more than 120 different tumor entities. 5751 tumor samples were analyzed on TMAs by immunohistochemistry (Hercept-Test, DAKO) and fluorescence in situ hybridization (PathVysion, Abbott-Vysis) under highly standardized conditions. HER-2 overexpression (score 2/3 þ ) and amplification occurred most often in breast cancers but was also seen in 18 other tumor entities including cancers of the urinary bladder (amplification in 14.3%, overexpression in 6.7%), stomach (8.3/4.9%), endometrium (6.6/6.8%), lung (2.8/3.1%) and ovary (2.3/ 1.2%). Remarkably, a strong association between overexpression and amplification was seen in all examined cancer entities. Trastuzumab therapy is highly efficient in HER-2 amplified breast cancer both in metastatic disease and as an adjuvant therapy. A variety of other tumor entities including frequent neoplasms and cancers with often limited therapeutic options have similar patterns of HER-2 alterations as observed in breast cancer (ie high overexpression due to high level gene amplification). -

Mrna In-Situ Hybridization Using Biotinylated Oligonucleotide Probes: Implications for the Diagnostic Laboratory* JULIANA G
ANNALS OF CLINICAL AND LABORATORY SCIENCE, Vol. 24, No. 4 Copyright © 1994, Institute for Clinical Science, Inc. mRNA In-situ Hybridization Using Biotinylated Oligonucleotide Probes: Implications for the Diagnostic Laboratory* JULIANA G. SZAKACS, M.D.t and SANDRA K. LIVINGSTON, HTL (ASCP)tt tDepartment of Pathology, tMolecular Histology Laboratory University of South Florida, College of Medicine Tampa, FL 33612 ABSTRACT It is now possible to detect low copy numbers of messenger ribonucleic acid (mRNA) while retaining good histologic morphology for the determi nation of specific gene expression in diseased tissues. This technology will allow the pathologist to provide important prognostic information about tumors (expression of oncogenes and growth factors), to identify the sub clones within the tumor which may be most likely to metastasize (expres sion of adhesion molecules and proteases) and to identify etiologic genetic aberrations (viral insertions). A technique for in-situ hybridization to mRNA has been developed for use in formalin fixed paraffin embedded tissues which is suitable for a hospital histology laboratory. Optimal con ditions for the procedure were determined by using a biotinylated poly (d)T oligonucleotide probe. Results were dependent on the tissue type, fixation time, condition of the tissue prior to fixation, and degree of diges tion before hybridization. The temperature and conditions of hybridization were optimized so that the poly d(T) control probe and the longer test probe could be run simultaneously. Streptavidin and avidin alkaline phos phatase detection systems were tested using levamisole to minimize back ground staining, and a biotin blocking agent to reduce reaction to renal tubular biotin. -

Seminars in Histology “In Situ Hybridization”
Seminars in Histology From basic principles to advanced histological techniques “In situ hybridization” DBM Histology Core Facility Dr. Diego Calabrese 07.09.2018 Introduction In Situ Hybridization (ISH) is a technique that allows for precise localization of nucleic acid within a histologic section. The underlying basis of ISH is that nucleic acids, if preserved adequately, can be detected through the application of a complementary strand of nucleic acid to which a reporter molecule is attached. The technique was originally developed by Pardue and Gall (1969) and (independently) by John et al. (1969). At this time radioisotopes were the only labels available for nucleic acids, and autoradiography was the only means of detecting hybridized sequences. Radioactive in situ techniques can detect low copy number mRNA molecules in individual cells (Harper et al., 1986). The terminal fluorochrome labeling procedure of RNA probes was developed by Bauman et al. (1980, 1984), in the same year the direct enzyme labeling procedure of nucleic acids was described by Renz and Kurz (1984). https://www.ncbi.nlm.nih.gov/probe/docs/techish/ Roche DIG Application Manual Why ISH Nucleic acid localization helps in addressing questions relative to: • Genomic DNA alterations • Gene amplification • Gene split • Gene translocation • Prenatal diagnostic • Gene expression • Expression in heterogeneous tissues • Co-expression • Pathogen presence and localization • Virus and bacteria localization In-situ hybridization systems ISH experiments can be classified based -

Fluorescence in Situ Hybridization
Neuropathology for the Neuroradiologist: PATHOLOGY REVIEW Fluorescence in Situ Hybridization F.J. Wippold II SUMMARY: Fluorescence in situ hybridization is a molecular cytogenetic technique that localizes A. Perry segments of DNA within tumor cells by using dyes that are visible with a fluorescent microscope. The technique has proved useful in typing a variety of tumors such as oligodendrogliomas and in under- standing the genetic forces driving oncogenesis. he field of molecular cytogenetics has opened an exciting quently named “chromosomes” or “stained bodies” by the Tavenue of discovery by using chromosome analysis to famous German anatomist Heinrich Wilhelm Gottfried identify clinically useful diagnostic, prognostic, and predictive von Waldeyer-Hartz (1836–1921).5 Waldeyer would also biomarkers. Neuropathologists have exploited this informa- describe such structures as the sweat glands of the eyelids tion with a number of molecular techniques, one of which is and the tonsillar ring, as well as propose the neuron theory known as fluorescence in situ hybridization (FISH) to subtype of the organization of the central nervous system, all of phenotypically similar neoplasms and to unlock the genetic which bear his name. Yet the significance of these tiny forces that drive tumor development. Appreciation of this tool threadlike structures and their role in heredity were not can enhance the interactions of the neuroradiologist with the appreciated until Thomas Hunt Morgan (1866–1945) pro- neuropathologist and deepen understanding of commonly posed that the packets of heredity, namely the genes, are occurring tumors beyond standard histologic recognition. stored on the chromosomes.6-8 Even the number of human The purpose of this report is to review the significance of FISH chromosomes was in doubt until one of the co-workers of in the context of diagnostic tumor pathology. -

Comparing Fluorescence in Situ Hybridization and Chromogenic In
Comparing Fluorescence In Situ Hybridization and Chromogenic In Situ Hybridization Methods to Determine the HER2/neu Status in Primary Breast Carcinoma using Tissue Microarray Kyeongmee Park, M.D., Ph.D., Jungyeon Kim, M.D., Ph.D., Sungjig Lim, M.D., Ph.D., Sehwan Han, M.D., Ph.D., Jung Young Lee, M.D., Ph.D. Departments of Pathology (PK, KJ, LS) and Surgery (HS), Inje University Sanggye Paik Hospital; and the Department of Pathology (LJ), College of Medicine, The Catholic University, Seoul, Korea prognostic implication because its amplification Identification of HER2/neu status is important for was associated with aggressive biologic features of predicting response to specific chemotherapy in the breast carcinoma. Integration of tissue microar- breast carcinoma. Chromogenic in situ hybridiza- ray technology enabled high-throughput determi- tion was performed using tissue microarray tech- nation of HER2/neu amplification profile with ra- nology on 188 primary breast carcinomas. To vali- pidity and accuracy in large cohorts of the breast date the reliability of novel chromogenic in situ carcinoma. hybridization technology, the results of chromo- genic in situ hybridization were correlated with the KEY WORDS: Breast carcinoma, Chromogenic in results of two-color fluorescence in situ hybridiza- situ hybridization, Fluorescence in situ hybridiza- tion done with the same tumors. On tissue microar- tion, HER2/neu, Tissue microarray. ray panels containing 188 breast carcinoma tissues, Mod Pathol 2003;16(9):937–943 fluorescence in situ hybridization and chromogenic in situ hybridization were conducted simulta- Among the numerous oncogenes and their prod- neously. HER2/neu amplification was detected in 46 ucts, HER2/neu is the most widely exploited one in tumors (24.5%) by fluorescence in situ hybridiza- clinical oncology. -
SABER Enables Highly Multiplexed and Amplified Detection of DNA And
bioRxiv preprint doi: https://doi.org/10.1101/401810; this version posted August 27, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. SABER enables highly multiplexed and amplified detection of DNA and RNA in cells and tissues Jocelyn Y. Kishi1;2;† Brian J. Beliveau1;2;† Sylvain W. Lapan3;† Emma R. West3 Allen Zhu1;2 Hiroshi M. Sasaki1;2 Sinem K. Saka1;2 Yu Wang1;2 Constance L. Cepko3;∗ Peng Yin1;2;∗ Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 021151 Department of Systems Biology, Harvard Medical School, Boston, MA 021152, Department of Genetics, Harvard Medical School, Boston, MA 021153, These authors contributed equally†, Correspondence∗: [email protected] (P.Y.), [email protected] (C.L.C.) SUMMARY of quantitative FISH signals. These strategies include the tar- geted deposition of detectable reactive molecules around the site Fluorescent in situ hybridization (FISH) reveals the abun- of probe hybridization,11–14 the targeted assembly of ‘branched’ dance and positioning of nucleic acid sequences in fixed sam- structures composed of DNA15–17 or locked nucleic acid (LNA) ples and can be combined with cell segmentation to produce molecules,18 the programmed in situ growth of detectable con- a powerful single cell gene expression assay. However, it re- catemers by enzymatic rolling circle amplification (RCA)19,20 mains difficult to label more than a few targets and to visu- or hybridization chain reaction (HCR),21–25 and the assembly of alize nucleic acids in environments such as thick tissue sam- topologically catenated DNA structures using serial rounds of ples using conventional FISH technologies. -
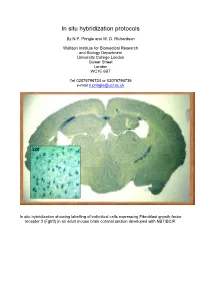
In Situ Hybridization Protocols
In situ hybridization protocols By N.P. Pringle and W. D. Richardson Wolfson Institute for Biomedical Research and Biology Department University College London Gower Street London WC1E 6BT Tel 02076796724 or 02076796736 e-mail [email protected] In situ hybridization showing labelling of individual cells expressing Fibroblast growth factor receptor 3 (Fgfr3) in an adult mouse brain coronal section developed with NBT/BCIP. Table of Contents OVERVIEW OF POTENTIAL PROBLEMS ............................................................................................................... 3 1. PROBE......................................................................................................................................................................... 3 2. TISSUE PREPARATION ................................................................................................................................................. 4 3. HYBRIDIZATION BUFFER ............................................................................................................................................ 4 4. CORRECT PH .............................................................................................................................................................. 4 MINIMAL PROTOCOL ................................................................................................................................................. 5 IN VITRO TRANSCRIPTION OF LABELED PROBES........................................................................................... -

(OLIG) As Molecular Markers for Human Glial Brain Tumors
Oligodendrocyte lineage genes (OLIG) as molecular markers for human glial brain tumors Q. Richard Lu*, John K. Park*†, Elizabeth Noll†, Jennifer A. Chan‡, John Alberta*, Dongin Yuk§, M. Garcia Alzamora*, David N. Louis¶, Charles D. Stiles*ʈ, David H. Rowitch§ʈ**, and Peter M. Black† Departments of *Cancer Biology and §Pediatric Oncology, the Program in Neuro-oncology, Dana–Farber͞Harvard Cancer Center, Dana–Farber Cancer Institute, 44 Binney Street, Boston, MA 02115; Department of †Neurosurgery and ‡Neuropathology, Brigham and Women’s Hospital, Boston, MA 02115; ¶Department of Pathology and Neurosurgical Service, Massachusetts General Hospital, Boston, MA 02129; and **Division of Newborn Medicine, Children’s Hospital, Boston, MA 02115 Communicated by Peter M. Howley, Harvard Medical School, Boston, MA, July 5, 2001 (received for review February 15, 2001) The most common primary tumors of the human brain are thought The absence of lineage-specific marker proteins for oligoden- to be of glial cell origin. However, glial cell neoplasms cannot be droglioma raises another question. Does oligodendroglioma fully classified by cellular morphology or with conventional mark- actually arise from oligodendrocytes or their progenitors? The ers for astrocytes, oligodendrocytes, or their progenitors. Recent term ‘‘oligodendroglioma’’ is empirical and reflects similarities insights into central nervous system tumorigenesis suggest that between the cellular morphology of well-differentiated tumors novel molecular markers might be found among factors that and nonneoplastic oligodendrocytes. Certain glial cell neo- have roles in glial development. Oligodendrocyte lineage genes plasms do express genes that are associated with undifferenti- (Olig1͞2) encode basic helix–loop–helix transcription factors. In the ated oligodendrocyte precursor cells (14).