STA-3900 Unsupervised Segmentation of Skin Lesions
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Electronic 3D Models Catalogue (On July 26, 2019)
Electronic 3D models Catalogue (on July 26, 2019) Acer 001 Acer Iconia Tab A510 002 Acer Liquid Z5 003 Acer Liquid S2 Red 004 Acer Liquid S2 Black 005 Acer Iconia Tab A3 White 006 Acer Iconia Tab A1-810 White 007 Acer Iconia W4 008 Acer Liquid E3 Black 009 Acer Liquid E3 Silver 010 Acer Iconia B1-720 Iron Gray 011 Acer Iconia B1-720 Red 012 Acer Iconia B1-720 White 013 Acer Liquid Z3 Rock Black 014 Acer Liquid Z3 Classic White 015 Acer Iconia One 7 B1-730 Black 016 Acer Iconia One 7 B1-730 Red 017 Acer Iconia One 7 B1-730 Yellow 018 Acer Iconia One 7 B1-730 Green 019 Acer Iconia One 7 B1-730 Pink 020 Acer Iconia One 7 B1-730 Orange 021 Acer Iconia One 7 B1-730 Purple 022 Acer Iconia One 7 B1-730 White 023 Acer Iconia One 7 B1-730 Blue 024 Acer Iconia One 7 B1-730 Cyan 025 Acer Aspire Switch 10 026 Acer Iconia Tab A1-810 Red 027 Acer Iconia Tab A1-810 Black 028 Acer Iconia A1-830 White 029 Acer Liquid Z4 White 030 Acer Liquid Z4 Black 031 Acer Liquid Z200 Essential White 032 Acer Liquid Z200 Titanium Black 033 Acer Liquid Z200 Fragrant Pink 034 Acer Liquid Z200 Sky Blue 035 Acer Liquid Z200 Sunshine Yellow 036 Acer Liquid Jade Black 037 Acer Liquid Jade Green 038 Acer Liquid Jade White 039 Acer Liquid Z500 Sandy Silver 040 Acer Liquid Z500 Aquamarine Green 041 Acer Liquid Z500 Titanium Black 042 Acer Iconia Tab 7 (A1-713) 043 Acer Iconia Tab 7 (A1-713HD) 044 Acer Liquid E700 Burgundy Red 045 Acer Liquid E700 Titan Black 046 Acer Iconia Tab 8 047 Acer Liquid X1 Graphite Black 048 Acer Liquid X1 Wine Red 049 Acer Iconia Tab 8 W 050 Acer -

Photography 101 Eric Kim a Gift to You
PHOTOGRAPHY 101 ERIC KIM A GIFT TO YOU Dear friend, I wanted to write you this book on how to take better photos. I know you started off with your iPhone, and started to show keen interest on making better images. I taught you the “rule of thirds” and other basics; but now it is time for you to take your photography to the next level. So consider this a handbook or a manual of sorts; for you only. I am also writing this book in hope that other people (similar to you, still relatively new in photography and wanting to learn more) will find it helpful as well. Always, Eric Dec 5, 2015 i 1 PHOTOGRAPHY EQUIPMENT WHAT CAMERA SHOUD I BUY? I wanted to have more manual controls. I wanted to do things like control my shutter speed, and also aperture (which is how much light the lens allows in, which also controls “depth-of-field”— which To start off, one of the first things on your mind is probably “what is how blurry the background is when you shoot (also called camera should I buy?” “bokeh”). Let me be honest with you— I regret spending so much time, effort, At the time, Flickr was still relatively new, Instagram wasn’t around, (and especially money) on all the gear and cameras and lenses I ac- Facebook was still in its infancy. The only way to share photos was cumulated over the years. They call it “Gear Acquisition Syndrome” either Flickr, photography forums (I was part of the “Black and White (G.A.S. -

Ricoh GR DIGITAL II User Manual
From environmental friendliness to environmental conservation and to environmental management Ricoh is aggressively promoting environment- friendly activities and also environment conservation activities to solve the great subject of management as one of the citizens on our precious earth. To reduce the environmental loads of digital cameras, Ricoh is also trying to solve the great subjects of “Saving energy by reducing power consumption” and “Reducing environment-affecting chemical substances contained in products”. If a problem arises First of all, see “Troubleshooting” (GP.211) in this manual. If the issues still persist, please contact a Ricoh office. Ricoh Offices RICOH COMPANY, LTD. 3-2-3, Shin-Yokohama Kouhoku-ku, Yokohama City, Kanagawa 222-8530, Japan http://www.ricoh.co.jp/r_dc RICOH AMERICAS CORPORATION 5 Dedrick Place, West Caldwell, New Jersey 07006, U.S.A. 1-800-22RICOH Camera User Guide http://www.ricoh-usa.com RICOH INTERNATIONAL B.V. (EPMMC) Oberrather Str. 6, 40472 Düsseldorf, GERMANY (innerhalb Deutschlands) 06331 268 438 (außerhalb Deutschlands) +49 6331 268 438 http://www.ricohpmmc.com The serial number of this product is given on the bottom face of the camera. RICOH UK LTD. (PMMC UK) (from within the UK) 02073 656 580 (from outside of the UK) +44 2073 656 580 RICOH FRANCE S.A.S. (PMMC FRANCE) (à partir de la France) 0800 91 4897 (en dehors de la France) +49 6331 268 439 RICOH ESPANA, S.A. (PMMC SPAIN) (desde España) 91 406 9148 Basic Operations (desde fuera de España) +34 91 406 9148 If you are using the camera for the first time, read this section. -
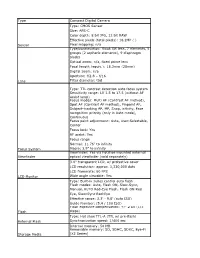
Type Compact Digital Camera Type: CMOS Sensor Size
Type Compact Digital Camera Type: CMOS Sensor Size: APS-C Color depth: 8 bit JPG, 12 bit RAW Effective pixels (total pixels): 16.2MP ( ) Sensor Pixel mapping: n/a Type/construction: Ricoh GR lens, 7 elements, 5 groups (2 aspheric elements), 9 diaphragm blades Optical zoom: n/a, fixed prime lens Focal length (equiv.): 18.3mm (28mm) Digital zoom: n/a Aperture: f/2.8 – f/16 Lens Filter diameter: tbd Type: TTL contrast detection auto focus system Sensitivity range: LV 1.5 to 17.5 (without AF assist lamp) Focus modes: Multi AF (Contrast AF method), Spot AF (Contrast AF method), Pinpoint AF, Subject-tracking AF, MF, Snap, infinity, Face recognition priority (only in Auto mode), Continuous Focus point adjustment: Auto, User-Selectable, Center Focus lock: Yes AF assist: Yes Focus range Normal: 11.76” to infinity Focus System Macro: 3.9” to infinity Viewfinder: Yes via hotshoe mounted external Viewfinder optical viewfinder (sold separately) 3.0" transparent LCD, w/ protective cover LCD resolution: approx. 1,230,000 dots LCD framerate: 60 FPS LCD Monitor Wide angle viewable: Yes Type: Built-in series control auto flash Flash modes: Auto, Flash ON, Slow-Sync, Manual, AUTO Red-Eye Flash, Flash ON Red Eye, Slow-Sync Red-Eye Effective range: 3.3’ - 9.8’ (auto ISO) Guide Number: (5.4 / 100 ISO) Flash exposure compensation: +/- 2 EV (1/3 Flash steps) Type: Hot shoe TTL-A (TTL w/ pre-flash) External Flash Synchronization speed: 1/400 sec Internal memory: 54 MB Removable memory: SD, SDHC, SDXC, Eye-Fi Storage Media (X2 Series) Ports: USB 2.0 hi-speed, AV/USB out, HDMI (Micro, Type D) Video out: NTSC, PAL, HD (HDMI supports HD Auto, 1080p, 720p, 480p) Interface(s) Microphone: Built-in monaural Power source: Rechargeable Li-Ion battery DB- 65 Recordable images: Li-Ion approx. -

Digital Image Ballistics from JPEG Quantization
TR2008-638, Dartmouth College, Computer Science 2 Methods Using the Flickr API, 1,000,000 images were down- loaded from Flickr.com, a popular photo-sharing Digital Image Ballistics website. Since we are interested in the JPEG quan- from JPEG Quantization: tization table employed by different cameras, it A Followup Study was necessary to eliminate any images that had been edited or altered by any photo-editing soft- ware. To this end, only images tagged as “orig- inal” by Flickr were downloaded. Images were Hany Farid then eliminated if they were not 3-channel color Department of Computer Science images, and further eliminated if they had incom- Dartmouth College plete metadata, inconsistent “modification” and Hanover NH 03755 “original” dates, or a “software” metadata tag sug- gesting that the image had been edited by a photo- editing software. This filtering eliminated 557,045 Abstract images, leaving 442,955 images. The camera make, model, resolution, and JPEG The lossy JPEG compression scheme quantization table were extracted from each of the employs a quantization table that con- 442,955 images’ metadata. In order to further elim- trols the amount of compression achieved. inate possible edited or altered images, only those Because different cameras typically em- entries with five or more images having the same ploy different tables, a comparison of paired make, model, resolution, and quantization an image’s quantization scheme to a table were retained. This filtering eliminated 105,329 database of known cameras affords a images, leaving 337,626 images for the final anal- simple technique for confirming or deny- ysis. -

New Tecnologies for Mobile Mapping
NEW TECNOLOGIES FOR MOBILE MAPPING Horea Bendea.a, Piero Boccardo.b, Sergio Dequal.a, Fabio Giulio Tonolo.b, Davide Marenchino.a a DITAG, Land Environment and Geo-Engineering Department of the Politecnico di Torino, C.so Duca degli Abruzzi 24, 10129 Turin, Italy – (horea.bendea, sergio.dequal, davide.marenchino)@polito.it b ITHACA, Information Technology for Humanitarian Assistance Cooperation and Action, Via Pier Carlo Boggio 61, 10138 Turin, Italy – (piero.boccardo, fabio.giuliotonolo) @ithaca.polito.it KEY WORDS: UAV, photogrammetry, payload, GPS/IMU, triangulation ABSTRACT: This paper deals with the development of a low cost UAV (Unmanned Aerial Vehicle) devoted to early impact phase in case of environmental disasters, based on geomatics techniques. “Pelican” is a low-cost UAV prototype equipped with a photogrammetric payload that will allows reconnaissance operations in remote areas and rapid mapping production. Different digital sensors installed in the payload allow to acquire high resolution frame images. Furthermore a GPS/INS unit will enable an automated navigation (except take-off and landing). The project is supported by ITHACA (Information Technology for Humanitarian Assistance, Cooperation and Action), an association founded by Politecnico di Torino and SiTI (Istituto Superiore sui Sistemi Territoriali per l’Innovazione) in cooperation with WFP (World Food Programme) and some private and public organisms, with the main goal to carry on operational and research activities in the field of geomatics for analysis, evaluation and mitigation of natural and manmade hazards. The main technical features of the UAV and the on-board payload are detailed described in the first part of the paper. Furthermore first results of stereopairs orientation, case studies and further developments are presented. -
AVAILABLE LIGHT Fotografieren Mit Wenig Licht
12 2014 MIT AKTUELLEN TEST Das Magazin BERICHTEN AUS DER 2,90€ oder gratis bei Ihrem RINGFOTO-Händler AVAILABLE LIGHT Fotografieren mit wenig Licht FOTOSCHULE Bildaufbau und Blickführung TEST & TECHNIK SLR gegen Spiegellose im Praxistest EDITORIAL Warte bis es dunkel wird ... In unseren Breiten ist es um diese Jahreszeit üblicherweise länger dunkel als hell und wer einen normalen Arbeitstag hat, geht morgens im Dunkeln aus dem Haus und kommt abends im Dunkeln zurück. Da bleibt für ein „lichtsensibles“ Hobby wie das Fotografieren nur das Wochenende, könnte man meinen. Von wegen. Digitale Kameras sind geradezu prädestiniert, mit ihnen im Dunkeln auf Fototour zu gehen. Wo man früher aufwendig niedrig- gegen hochempfindliche Filme tauschen musste, genügt heute ein einfacher Dreh an der ISO-Emp- Claudia Endres findlichkeit der Kamera. Und Multishot-Verfahren wie HDR Leiterin Marketing / Vertrieb der RINGFOTO-Gruppe eröffnen uns Möglichkeiten, von denen wir vor zehn oder fünfzehn Jahren nicht einmal geträumt hätten. Unser Beitrag zum Fotografieren bei wenig Licht gibt Ihnen viele Tipps, wie Sie bei Nacht und in der Dämmerung zu guten Bildern kommen. Und ich muss sagen: Nachdem ich das eine oder andere selbst ausprobiert habe, warte ich manchmal schon sehnsüchtig darauf, dass es endlich dunkel wird ... Viel Spaß beim Lesen wünscht Ihnen Titel: © andreiuc88 – shutterstock.com 03 INHALT 14 PRAXIS Tipps und Tricks für die Fotografie bei wenig Licht 30 SLR GEGEN SPIEGELLOSE Die Canon EOS 7D und die Olympus OM-D E-M1 im großen Praxistest 04 Inhalt EDITORIAL 3 Warte bis es dunkel wird ... NEWS 6 Trends und Neuheiten BUCHTIPP 8 Fremdes Volk ZUBEHÖR 10 Rahmen von Peter Hadley EVENTKALENDER 11 Ausstellungen ZUBEHÖR 12 Eddycam Kameragurte 22 PRAXIS 14 PRAXIS Fotografie bei wenig Licht Bildaufbau und Blickführung PRAXIS 22 Fotoschule 10 – Bildgestaltung AKTIONSPRODUKT 28 Canon IXUS 150 TESTBERICHT 30 Canon EOS 7D vs. -
Get Raw Working with Your Camera
RAW yoUR COMPLETE GET YOUR RAW RAW COMPATIBILITY GUIDE FILES worKING Now REFERENCE tablE PRINT-OUT & KEEP PDF GUIDE + PRINT-OUT & KEEP PDF GUIDE + PRINT-OUT GET YOUR RAWS GET RAW WORKING WITH YOUR CAMERA WORKING NOW! This guide is designed EVERY NEW CAMERA MODEL that hits the NEF from a Nikon D200 camera, for example, is you upgrade or update it. This Making Photoshop or Elements shelves has a unique type of RAW format, and completely different to a NEF from a D300, and compatibility problem accounts for about compatible with your camera’s to get RAWs from that means you’ll almost certainly need to a CR2 from a Canon 450D is nothing like the 90 percent of the RAW questions we RAWs means downloading the latest upgrade or at least update your RAW CR2 from a 550D. It doesn’t matter if the file receive, and though it seems confusing at update for Adobe Camera Raw. your camera working conversion software if you invest in a new extension is the same – what’s important is first, it’s easy to swallow if you simply Unless you have the latest version of with your version of camera. There are different proprietary RAW that your camera model is supported by your remember that every camera has its own Photoshop or Elements, though, this may formats from each maker, such as NEF from RAW software, and that’s down to how recent bespoke RAW format, and if your camera also mean upgrading the software, too, Photoshop... Nikon, CR2 from Canon, PEF from Pentax, ORF the software is compared to your camera. -

Public Auction
16 Raffles Quay #30-01 Hong Leong Building Singapore 048581 Tel: 6228 7380/ 6228 7302 Fax: 6221 9305 http://www.knightfrank.com.sg/singapore-auction-property PUBLIC AUCTION BY ORDER OF SINGAPORE POLICE FORCE KNIGHT FRANK PTE LTD Will Sell by Public Auction Brass Cartridges, Outboard engine, Vehicles (Saloon cars & Motor- Cycles) Confiscated & Unclaimed items, Jewellery Branded watches, IT & Info-comm equipments, Office & home appliances, Bicycles, Air-cons and Digital Cameras (Viewing Venues set at Various Locations) Auction Venue : Village Hotel Bugis (formerly Landmark Hotel) (No viewing @ venue) Quartz 1 Room, Level 6 390 Victoria Street, Singapore 188061 Date & Time of Auction : 4 March 2015, Wednesday Starts at 11.00am Viewing Venues : Please refer to Auction catalogues for the viewing locations of specific items/ vehicles Viewing Dates & Time : 2 & 3 March 2015, Monday & Tuesday 10am to 12noon and 2pm to 4pm Featured Items : Patek Philippe, Mont Blanc, Swarovski, Gucci, Braun Buffel, Nike, Samsung Galaxy, DSLR cameras and many more Important Notes on : Prospective bidders are required to present their Brass Cartridges valid licence upon registration to be able to bid for the handling and disposing of empty brass cartridges. For Sale Catalogues, Conditions of Sale and for further information, please contact Auctioneer. GENERAL CONDITIONS OF SALE 1 The sale will be by way of public auction. 2. The highest bidder shall be the buyer, and if any dispute arises among the bidders, the lot in dispute shall, at the Auctioneer's discretion, either be immediately put up again and resold or the Auctioneer may decide the dispute. The Authority reserves the right to alter, vary or withdraw any lot or lots before or during the sale. -

Tropical Peatland Burn Depth and Combustion Heterogeneity Assessed Using UAV Photogrammetry and Airborne Lidar
remote sensing Article Tropical Peatland Burn Depth and Combustion Heterogeneity Assessed Using UAV Photogrammetry and Airborne LiDAR Jake E. Simpson 1,*, Martin J. Wooster 1,2, Thomas E. L. Smith 1, Mandar Trivedi 3, Ronald R. E. Vernimmen 4, Rahman Dedi 5, Mulya Shakti 5 and Yoan Dinata 5 1 Department of Geography, King’s College London, London WC2R 2LS, UK; [email protected] (M.J.W.); [email protected] (T.E.L.S.) 2 National Centre for Earth Observation (NCEO), Natural Environment Research Council, Leicester LE1 7RH, UK 3 Zoological Society of London, London NW1 4RY, UK; [email protected] 4 Deltares, Boussinesqweg 1, 2629 HV Delft, The Netherlands; [email protected] 5 Zoological Society of London Indonesia, Kota Jambi 36124, Sumatra, Indonesia; [email protected] (R.D.); [email protected] (M.S.); [email protected] (Y.D.) * Correspondence: [email protected] Academic Editors: Krishna Prasad Vadrevu, Rama Nemani, Chris Justice, Garik Gutman, Ioannis Gitas and Prasad S. Thenkabail Received: 4 October 2016; Accepted: 29 November 2016; Published: 6 December 2016 Abstract: We provide the first assessment of tropical peatland depth of burn (DoB) using structure from motion (SfM) photogrammetry, applied to imagery collected using a low-cost, low-altitude unmanned aerial vehicle (UAV) system operated over a 5.2 ha tropical peatland in Jambi Province on Sumatra, Indonesia. Tropical peat soils are the result of thousands of years of dead biomass accumulation, and when burned are globally significant net sources of carbon emissions. The El Niño year of 2015 saw huge areas of Indonesia affected by tropical peatland fires, more so than any year since 1997. -

Supported Cameras Luminar 2
"AgfaPhoto DC-833m", "Alcatel 5035D", "Apple iPad Pro", "Apple iPhone SE", "Apple iPhone 6s", "Apple iPhone 6 plus", "Apple iPhone 7", "Apple iPhone 7 plus", "Apple iPhone 8”, "Apple iPhone 8 plus”, "Apple iPhone X”, "Apple QuickTake 100", "Apple QuickTake 150", "Apple QuickTake 200", "ARRIRAW format", "AVT F-080C", "AVT F-145C", "AVT F-201C", "AVT F-510C", "AVT F-810C", "Baumer TXG14", "BlackMagic Cinema Camera", "BlackMagic Micro Cinema Camera", "BlackMagic Pocket Cinema Camera", "BlackMagic Production Camera 4k", "BlackMagic URSA", "BlackMagic URSA Mini 4k", "BlackMagic URSA Mini 4.6k", "BlackMagic URSA Mini Pro 4.6k", "Canon PowerShot 600", "Canon PowerShot A5", "Canon PowerShot A5 Zoom", "Canon PowerShot A50", "Canon PowerShot A410", "Canon PowerShot A460", "Canon PowerShot A470", "Canon PowerShot A530", "Canon PowerShot A540", "Canon PowerShot A550", "Canon PowerShot A570", "Canon PowerShot A590", "Canon PowerShot A610", "Canon PowerShot A620", "Canon PowerShot A630", "Canon PowerShot A640", "Canon PowerShot A650", "Canon PowerShot A710 IS", "Canon PowerShot A720 IS", "Canon PowerShot A3300 IS", "Canon PowerShot D10", "Canon PowerShot ELPH 130 IS", "Canon PowerShot ELPH 160 IS", "Canon PowerShot Pro70", "Canon PowerShot Pro90 IS", "Canon PowerShot Pro1", "Canon PowerShot G1", "Canon PowerShot G1 X", "Canon PowerShot G1 X Mark II", "Canon PowerShot G1 X Mark III”, "Canon PowerShot G2", "Canon PowerShot G3", "Canon PowerShot G3 X", "Canon PowerShot G5", "Canon PowerShot G5 X", "Canon PowerShot G6", "Canon PowerShot G7", "Canon PowerShot -

Capture One 6.3.5 Release Notes
Capture One 6.3.1 Release Notes Introduction Capture One 6 is a raw converter and workflow software which enables photographers to reduce the time and effort required to deliver stunning ready-to-use images with excellent color and detail. Capture One 6 is designed to create the best image quality on the market and holds a series of easy-to-use tools created to match the professional photographer’s daily workflow. Capture One 6 is made by Phase One, the World’s leading manufacturer of high-end digital camera systems, in collaboration with the World’s leading professional photographers. Capture One 6 comes in three versions: Express, Pro and DB. Capture One 6.3.1 is a free update to existing owners of Capture One 6. Release Notes Phase One March 2012 Page 1 of 16 Capture One 6.3.5 Release Notes Capture One 6 is a raw converter and workflow software which enables photographers to reduce the time and effort required to deliver stunning ready-to-use images with excellent color and detail. Capture One 6 is designed to create the best image quality on the market and holds a series of easy-to-use tools created to match the professional photographer’s daily workflow. Capture One 6 is made by Phase One, the World’s leading manufacturer of high-end digital camera systems, in collaboration with the World’s leading professional photographers. Capture One 6 comes in three versions: Express, Pro and DB. Capture One 6.3.5 is a free update to existing owners of Capture One 6.