Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

RI-Mediated Mast Cell Activation Ε of Fc Tetraspanin CD151 Is A
Tetraspanin CD151 Is a Negative Regulator of Fc εRI-Mediated Mast Cell Activation Hiam Abdala-Valencia, Paul J. Bryce, Robert P. Schleimer, Joshua B. Wechsler, Lucas F. Loffredo, Joan M. Cook-Mills, This information is current as Chia-Lin Hsu and Sergejs Berdnikovs of September 26, 2021. J Immunol 2015; 195:1377-1387; Prepublished online 1 July 2015; doi: 10.4049/jimmunol.1302874 http://www.jimmunol.org/content/195/4/1377 Downloaded from Supplementary http://www.jimmunol.org/content/suppl/2015/07/01/jimmunol.130287 Material 4.DCSupplemental http://www.jimmunol.org/ References This article cites 63 articles, 28 of which you can access for free at: http://www.jimmunol.org/content/195/4/1377.full#ref-list-1 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision by guest on September 26, 2021 • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2015 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology Tetraspanin CD151 Is a Negative Regulator of Fc«RI-Mediated Mast Cell Activation Hiam Abdala-Valencia,* Paul J. -

Fine Tuning by Human Cd1e of Lipid-Specific Immune Responses
Fine tuning by human CD1e of lipid-specific immune responses Federica Facciottia, Marco Cavallaria, Catherine Angénieuxb, Luis F. Garcia-Allesc, François Signorino-Gelob, Lena Angmana, Martine Gilleronc, Jacques Prandic, Germain Puzoc, Luigi Panzad, Chengfeng Xiae, Peng George Wangf, Paolo Dellabonag, Giulia Casoratig, Steven A. Porcellih, Henri de la Salleb, Lucia Moria,i,1, and Gennaro De Liberoa,1 aExperimental Immunology, Department of Biomedicine, University Hospital Basel, 4031 Basel, Switzerland; bInstitut National de la Santé et de la Recherche Médicale, Unité Mixte de Recherche S725, Biology of Human Dendritic Cells, Université de Strasbourg and Etablissement Français du Sang-Alsace, 67065 Strasbourg, France; cCentre National de la Recherche Scientifique, Institut de Pharmacologie et de Biologie Structurale, 31077 Toulouse, France; dDepartment of Chemistry, Food, Pharmaceuticals, and Pharmacology, Università del Piemonte Orientale, 28100 Novara, Italy; eState Key Laboratory of Phytochemistry and Plant Resources in West China, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan 650204, China; fDepartment of Biochemistry and Chemistry, The Ohio State University, Columbus, OH 43210; gExperimental Immunology Unit, Division of Immunology, Transplantation, and Infectious Diseases, Dipartimento di Biotecnologie (DIBIT), San Raffaele Scientific Institute, 20132 Milano, Italy; hAlbert Einstein College of Medicine, Bronx, NY 10461; and iSingapore Immunology Network, Agency for Science Technology and Research, Biopolis, Singapore 138648 Edited by Peter Cresswell, Yale University School of Medicine, New Haven, CT, and approved July 20, 2011 (received for review June 5, 2011) CD1e is a member of the CD1 family that participates in lipid an- molecules traffic to early recycling endosomes and, to a lesser tigen presentation without interacting with the T-cell receptor. -

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -

Supporting Online Material
1 2 3 4 5 6 7 Supplementary Information for 8 9 Fractalkine-induced microglial vasoregulation occurs within the retina and is altered early in diabetic 10 retinopathy 11 12 *Samuel A. Mills, *Andrew I. Jobling, *Michael A. Dixon, Bang V. Bui, Kirstan A. Vessey, Joanna A. Phipps, 13 Ursula Greferath, Gene Venables, Vickie H.Y. Wong, Connie H.Y. Wong, Zheng He, Flora Hui, James C. 14 Young, Josh Tonc, Elena Ivanova, Botir T. Sagdullaev, Erica L. Fletcher 15 * Joint first authors 16 17 Corresponding author: 18 Prof. Erica L. Fletcher. Department of Anatomy & Neuroscience. The University of Melbourne, Grattan St, 19 Parkville 3010, Victoria, Australia. 20 Email: [email protected] ; Tel: +61-3-8344-3218; Fax: +61-3-9347-5219 21 22 This PDF file includes: 23 24 Supplementary text 25 Figures S1 to S10 26 Tables S1 to S7 27 Legends for Movies S1 to S2 28 SI References 29 30 Other supplementary materials for this manuscript include the following: 31 32 Movies S1 to S2 33 34 35 36 1 1 Supplementary Information Text 2 Materials and Methods 3 Microglial process movement on retinal vessels 4 Dark agouti rats were anaesthetized, injected intraperitoneally with rhodamine B (Sigma-Aldrich) to label blood 5 vessels and retinal explants established as described in the main text. Retinal microglia were labelled with Iba-1 6 and imaging performed on an inverted confocal microscope (Leica SP5). Baseline images were taken for 10 7 minutes, followed by the addition of PBS (10 minutes) and then either fractalkine or fractalkine + candesartan 8 (10 minutes) using concentrations outlined in the main text. -

Transcriptional Control of Tissue-Resident Memory T Cell Generation
Transcriptional control of tissue-resident memory T cell generation Filip Cvetkovski Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Graduate School of Arts and Sciences COLUMBIA UNIVERSITY 2019 © 2019 Filip Cvetkovski All rights reserved ABSTRACT Transcriptional control of tissue-resident memory T cell generation Filip Cvetkovski Tissue-resident memory T cells (TRM) are a non-circulating subset of memory that are maintained at sites of pathogen entry and mediate optimal protection against reinfection. Lung TRM can be generated in response to respiratory infection or vaccination, however, the molecular pathways involved in CD4+TRM establishment have not been defined. Here, we performed transcriptional profiling of influenza-specific lung CD4+TRM following influenza infection to identify pathways implicated in CD4+TRM generation and homeostasis. Lung CD4+TRM displayed a unique transcriptional profile distinct from spleen memory, including up-regulation of a gene network induced by the transcription factor IRF4, a known regulator of effector T cell differentiation. In addition, the gene expression profile of lung CD4+TRM was enriched in gene sets previously described in tissue-resident regulatory T cells. Up-regulation of immunomodulatory molecules such as CTLA-4, PD-1, and ICOS, suggested a potential regulatory role for CD4+TRM in tissues. Using loss-of-function genetic experiments in mice, we demonstrate that IRF4 is required for the generation of lung-localized pathogen-specific effector CD4+T cells during acute influenza infection. Influenza-specific IRF4−/− T cells failed to fully express CD44, and maintained high levels of CD62L compared to wild type, suggesting a defect in complete differentiation into lung-tropic effector T cells. -

Uva-DARE (Digital Academic Repository)
UvA-DARE (Digital Academic Repository) Balancing effector lymphocyte formation via CD27-CD70 interactions Arens, R. Publication date 2003 Link to publication Citation for published version (APA): Arens, R. (2003). Balancing effector lymphocyte formation via CD27-CD70 interactions. General rights It is not permitted to download or to forward/distribute the text or part of it without the consent of the author(s) and/or copyright holder(s), other than for strictly personal, individual use, unless the work is under an open content license (like Creative Commons). Disclaimer/Complaints regulations If you believe that digital publication of certain material infringes any of your rights or (privacy) interests, please let the Library know, stating your reasons. In case of a legitimate complaint, the Library will make the material inaccessible and/or remove it from the website. Please Ask the Library: https://uba.uva.nl/en/contact, or a letter to: Library of the University of Amsterdam, Secretariat, Singel 425, 1012 WP Amsterdam, The Netherlands. You will be contacted as soon as possible. UvA-DARE is a service provided by the library of the University of Amsterdam (https://dare.uva.nl) Download date:27 Sep 2021 Chapter 3 Constitutive CD27/CD70 interaction induces expansion of effector-type T cells and results in IFNy-mediated B cell depletion Ramon Arens*, Kiki Tesselaar*, Paul A. Baars, Gijs M.W. van Schijndel, Jenny Hendriks, Steven T. Pals, Paul Krimpenfort, Jannie Borst, Marinus H.J. van Oers, and René A.W. van Lier 'These authors contributed equally to this work Immunity 15, 801-812 (2001) Chapter 3 Constitutive CD27/CD70 interaction induces expansion of effector-type T cells and results in IFNy-mediated B cell depletion Ramon Arens123#, Kiki Tesselaar23", Paul A. -

SUPPLEMENTARY TABLES and FIGURE LEGENDS Supplementary
SUPPLEMENTARY TABLES AND FIGURE LEGENDS Supplementary Figure 1. Quantitation of MYC levels in vivo and in vitro. a) MYC levels in cell lines 6814, 6816, 5720, 966, and 6780 (corresponding to first half of Figure 1a in main text). MYC is normalized to tubulin. b) MYC quantitations (normalized to tubulin) for cell lines Daudi, Raji, Jujoye, KRA, KRB, GM, and 6780 corresponding to second half of Figure 1a. c) In vivo MYC quantitations, for mice treated with 0-0.5 ug/ml doxycycline in their drinking water. MYC is normalized to tubulin. d) Quantitation of changing MYC levels during in vitro titration, normalized to tubulin. e) Levels of Odc (normalized to tubulin) follow MYC levels in titration series. Supplementary Figure 2. Evaluation of doxycycline concentration in the plasma of mice treated with doxycycline in their drinking water. Luciferase expressing CHO cells (Tet- off) (Clonethech Inc) that is responsive to doxycycline by turning off luciferase expression was treated with different concentrations of doxycycline in culture. A standard curve (blue line) correlating luciferase activity (y-axis) with treatment of doxycycline (x- axis) was generated for the CHO cell in culture. Plasma from mice treated with different concentrations of doxycycline in their drinking water was separated and added to the media of the CHO cells. Luciferase activity was measured and plotted on the standard curve (see legend box). The actual concentration of doxycycline in the plasma was extrapolated for the luciferase activity measured. The doxycycline concentration 0.2 ng/ml measured in the plasma of mice correlates with 0.05 μg/ml doxycycline treatment in the drinking water of mice, the in vivo threshold for tumor regression. -

Expression of the Tetraspanins CD9, CD37, CD63, and CD151 in Merkel Cell Carcinoma: Strong Evidence for a Posttranscriptional Fine-Tuning of CD9 Gene Expression
Modern Pathology (2010) 23, 751–762 & 2010 USCAP, Inc. All rights reserved 0893-3952/10 $32.00 751 Expression of the tetraspanins CD9, CD37, CD63, and CD151 in Merkel cell carcinoma: strong evidence for a posttranscriptional fine-tuning of CD9 gene expression Markus Woegerbauer1, Dietmar Thurnher1, Roland Houben2, Johannes Pammer3, Philipp Kloimstein1, Gregor Heiduschka1, Peter Petzelbauer4 and Boban M Erovic1 1Department of Otorhinolaryngology, Head and Neck Surgery, Medical University of Vienna, Vienna, Austria; 2Department of Dermatology, Medical University of Wuerzburg, Germany; 3Department of Clinical Pathology, Medical University of Vienna, Vienna, Austria and 4Department of Dermatology, Medical University of Vienna, Vienna, Austria Tetraspanins including CD9, CD37, CD63, and CD151 are linked to cellular adhesion, cell differentiation, migration, carcinogenesis, and tumor progression. The aim of the study was to detect, quantify, and evaluate the prognostic value of these tetraspanins in Merkel cell carcinoma and to study the regulation of CD9 mRNA expression in Merkel cell carcinoma cell lines in detail. Immunohistochemical staining of 28 Merkel cell carcinoma specimens from 25 patients showed a significant correlation of CD9 (P ¼ 0.03) and CD151 (P ¼ 0.043) expression to overall survival. CD9 and CD63 expression correlated significantly to patients’ disease-free interval (P ¼ 0.017 and P ¼ 0.058). Of primary Merkel cell carcinoma tumors, 42% were CD9 positive in contrast to only 21% of the subcutaneous in-transit metastases. Characterization of the 50 untranslated region (UTR) of the CD9 mRNA from two cultured Merkel cell carcinoma cell lines revealed the presence of two major RNA species differing only in the length of their 50 termini (183 versus 102 nucleotides). -
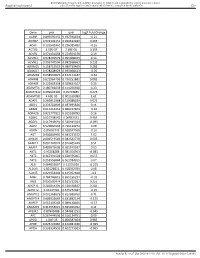
Gene Pval Qval Log2 Fold Change AAMP 0.895690332 0.952598834
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) Gut Gene pval qval Log2 Fold Change AAMP 0.895690332 0.952598834 -0.21 ABI3BP 0.002302151 0.020612283 0.465 ACHE 0.103542461 0.296385483 -0.16 ACTG2 2.99E-07 7.68E-05 3.195 ACVR1 0.071431098 0.224504378 0.19 ACVR1C 0.978209579 0.995008423 0.14 ACVRL1 0.006747504 0.042938663 0.235 ADAM15 0.158715519 0.380719469 0.285 ADAM17 0.978208929 0.995008423 -0.05 ADAM28 0.038932876 0.152174187 -0.62 ADAM8 0.622964796 0.790251882 0.085 ADAM9 0.122003358 0.329623107 0.25 ADAMTS1 0.180766659 0.414256926 0.23 ADAMTS12 0.009902195 0.05703885 0.425 ADAMTS8 4.60E-05 0.001169089 1.61 ADAP1 0.269811968 0.519388039 0.075 ADD1 0.233702809 0.487695826 0.11 ADM2 0.012213453 0.066227879 -0.36 ADRA2B 0.822777921 0.915518785 0.16 AEBP1 0.010738542 0.06035531 0.465 AGGF1 0.117946691 0.320915024 -0.095 AGR2 0.529860903 0.736120272 0.08 AGRN 0.85693743 0.928047568 -0.16 AGT 0.006849995 0.043233572 1.02 AHNAK 0.006519543 0.042542779 0.605 AKAP12 0.001747074 0.016405449 0.51 AKAP2 0.409929603 0.665919397 0.05 AKT1 0.95208288 0.985354963 -0.085 AKT2 0.367391504 0.620376005 0.055 AKT3 0.253556844 0.501934205 0.07 ALB 0.064833867 0.21195036 -0.315 ALDOA 0.83128831 0.918352939 0.08 ALOX5 0.029954404 0.125352668 -0.3 AMH 0.784746815 0.895196237 -0.03 ANG 0.050500474 0.181732067 0.255 ANGPT1 0.281853305 0.538528647 0.285 ANGPT2 0.43147281 0.675272487 -0.15 ANGPTL2 0.001368876 0.013688762 0.71 ANGPTL4 0.686032669 0.831882134 -0.175 ANPEP 0.019103243 0.089148466 -0.57 ANXA2P2 0.412553021 0.665966092 0.11 AP1M2 0.87843088 0.944681253 -0.045 APC 0.267444505 0.516134751 0.09 APOD 1.04E-05 0.000587404 0.985 APOE 0.023722987 0.104981036 -0.395 APOH 0.336334555 0.602273505 -0.065 Sundar R, et al. -

Increased Expression of CD154 and FAS in SLE Patients’ Lymphocytes Maria Elena Manea, Ruediger B
Increased expression of CD154 and FAS in SLE patients’ lymphocytes Maria Elena Manea, Ruediger B. Mueller, Doru Dejica, Ahmed Sheriff, Georg Schett, Martin Herrmann, Peter Kern To cite this version: Maria Elena Manea, Ruediger B. Mueller, Doru Dejica, Ahmed Sheriff, Georg Schett, et al.. Increased expression of CD154 and FAS in SLE patients’ lymphocytes. Rheumatology International, Springer Verlag, 2009, 30 (2), pp.181-185. 10.1007/s00296-009-0933-4. hal-00568285 HAL Id: hal-00568285 https://hal.archives-ouvertes.fr/hal-00568285 Submitted on 23 Feb 2011 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Increased expression of CD154 and FAS in SLE patients’ lymphocytes Maria Elena Manea1‡, MD, Ruediger B. Mueller2,3‡, MD, Doru Dejica1, PhD, Ahmed Sheriff2, PhD, Georg Schett2, MD, Martin Herrmann2, PhD, Peter Kern4, MD 1 Department of Immunopathology. “Iuliu Hatieganu" University of Medicine and Pharmacy, Str Croitorilor no 19-21, 3400 Cluj-Napoca, Romania. 2 Department for Internal Medicine 3 and Institute for Clinical Immunology, University of Erlangen-Nürnberg, Germany 3 Departement of Rheumatologie, Kantonsspital St. Gallen, Switzerland 4 Franz von Prümmer Klinik, Bahnhofstraße 16, 97769 Bad Brückenau, Germany ‡ both authors equally contributed to the work Address correspondence and reprint requests to: Ruediger B. -

Cell-Expressed CD154 in Germinal Centers Expression, Regulation
Expression, Regulation, and Function of B Cell-Expressed CD154 in Germinal Centers Amrie C. Grammer, Richard D. McFarland, Jonathan Heaney, Bonnie F. Darnell and Peter E. Lipsky This information is current as of September 25, 2021. J Immunol 1999; 163:4150-4159; ; http://www.jimmunol.org/content/163/8/4150 Downloaded from References This article cites 74 articles, 33 of which you can access for free at: http://www.jimmunol.org/content/163/8/4150.full#ref-list-1 Why The JI? Submit online. http://www.jimmunol.org/ • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average by guest on September 25, 2021 Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 1999 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. Expression, Regulation, and Function of B Cell-Expressed CD154 in Germinal Centers1 Amrie C. Grammer,* Richard D. McFarland,† Jonathan Heaney,* Bonnie F. Darnell,† and Peter E. Lipsky2* Activated B cells and T cells express CD154/CD40 ligand in vitro. The in vivo expression and function of B cell CD154 remain unclear and therefore were examined. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like,