Digital Atlas That Maps Signalling Pathways On
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

GLAAD Media Institute Began to Track LGBTQ Characters Who Have a Disability
Studio Responsibility IndexDeadline 2021 STUDIO RESPONSIBILITY INDEX 2021 From the desk of the President & CEO, Sarah Kate Ellis In 2013, GLAAD created the Studio Responsibility Index theatrical release windows and studios are testing different (SRI) to track lesbian, gay, bisexual, transgender, and release models and patterns. queer (LGBTQ) inclusion in major studio films and to drive We know for sure the immense power of the theatrical acceptance and meaningful LGBTQ inclusion. To date, experience. Data proves that audiences crave the return we’ve seen and felt the great impact our TV research has to theaters for that communal experience after more than had and its continued impact, driving creators and industry a year of isolation. Nielsen reports that 63 percent of executives to do more and better. After several years of Americans say they are “very or somewhat” eager to go issuing this study, progress presented itself with the release to a movie theater as soon as possible within three months of outstanding movies like Love, Simon, Blockers, and of COVID restrictions being lifted. May polling from movie Rocketman hitting big screens in recent years, and we remain ticket company Fandango found that 96% of 4,000 users hopeful with the announcements of upcoming queer-inclusive surveyed plan to see “multiple movies” in theaters this movies originally set for theatrical distribution in 2020 and summer with 87% listing “going to the movies” as the top beyond. But no one could have predicted the impact of the slot in their summer plans. And, an April poll from Morning COVID-19 global pandemic, and the ways it would uniquely Consult/The Hollywood Reporter found that over 50 percent disrupt and halt the theatrical distribution business these past of respondents would likely purchase a film ticket within a sixteen months. -

Open Source Used in Cisco Prime Access Registrar 7.3
Open Source Used In Cisco Prime Access Registrar 7.3 Cisco Systems, Inc. www.cisco.com Cisco has more than 200 offices worldwide. Addresses, phone numbers, and fax numbers are listed on the Cisco website at www.cisco.com/go/offices. Text Part Number: 78EE117C99-148346687 Open Source Used In Cisco Prime Access Registrar 7.3 1 This document contains licenses and notices for open source software used in this product. With respect to the free/open source software listed in this document, if you have any questions or wish to receive a copy of any source code to which you may be entitled under the applicable free/open source license(s) (such as the GNU Lesser/General Public License), please contact us at [email protected]. In your requests please include the following reference number 78EE117C99-148346687 Contents 1.1 ace-5.5 5.5 1.1.1 Available under license 1.2 ajaxanywhere 1.2.1 1.2.1 Available under license 1.3 ant 1.7.1 1.3.1 Available under license 1.4 Apache Commons Logging 1.1.1 1.4.1 Available under license 1.5 Apache Tomcat 8.5.16 1.5.1 Available under license 1.6 axis 1.4 1.6.1 Available under license 1.7 Axis C++ 1.6b 1.7.1 Available under license 1.8 Bean Validation API 1.1.0.Final 1.8.1 Available under license 1.9 c-ares 1.10.0 1.9.1 Available under license 1.10 CGLIB 2.2 1.10.1 Available under license 1.11 cglib 2.2.0-b21 :repackaged as a module 1.11.1 Available under license 1.12 cglib 2.1.88 1.12.1 Available under license 1.13 ciscosafec-3.0.0 3.0.0 1.13.1 Available under license Open Source Used In Cisco -
Unravel Data Systems Version 4.5
UNRAVEL DATA SYSTEMS VERSION 4.5 Component name Component version name License names jQuery 1.8.2 MIT License Apache Tomcat 5.5.23 Apache License 2.0 Tachyon Project POM 0.8.2 Apache License 2.0 Apache Directory LDAP API Model 1.0.0-M20 Apache License 2.0 apache/incubator-heron 0.16.5.1 Apache License 2.0 Maven Plugin API 3.0.4 Apache License 2.0 ApacheDS Authentication Interceptor 2.0.0-M15 Apache License 2.0 Apache Directory LDAP API Extras ACI 1.0.0-M20 Apache License 2.0 Apache HttpComponents Core 4.3.3 Apache License 2.0 Spark Project Tags 2.0.0-preview Apache License 2.0 Curator Testing 3.3.0 Apache License 2.0 Apache HttpComponents Core 4.4.5 Apache License 2.0 Apache Commons Daemon 1.0.15 Apache License 2.0 classworlds 2.4 Apache License 2.0 abego TreeLayout Core 1.0.1 BSD 3-clause "New" or "Revised" License jackson-core 2.8.6 Apache License 2.0 Lucene Join 6.6.1 Apache License 2.0 Apache Commons CLI 1.3-cloudera-pre-r1439998 Apache License 2.0 hive-apache 0.5 Apache License 2.0 scala-parser-combinators 1.0.4 BSD 3-clause "New" or "Revised" License com.springsource.javax.xml.bind 2.1.7 Common Development and Distribution License 1.0 SnakeYAML 1.15 Apache License 2.0 JUnit 4.12 Common Public License 1.0 ApacheDS Protocol Kerberos 2.0.0-M12 Apache License 2.0 Apache Groovy 2.4.6 Apache License 2.0 JGraphT - Core 1.2.0 (GNU Lesser General Public License v2.1 or later AND Eclipse Public License 1.0) chill-java 0.5.0 Apache License 2.0 Apache Commons Logging 1.2 Apache License 2.0 OpenCensus 0.12.3 Apache License 2.0 ApacheDS Protocol -

Progeny Upgrade Guide
Progeny Upgrade Guide Progeny Upgrade Guide Page | 1 Contents Getting Started .............................................................................................................................................. 3 A note about Progeny Downloads and Updates .............................................................................. 3 Progeny Desktop Client Update/Upgrade .................................................................................................... 3 Before Updating/Upgrading the Progeny Desktop Client ............................................................... 3 Upgrading the Progeny Desktop between major versions (Ex. 9 to 10) .......................................... 4 Updating the Progeny Desktop between minor versions (Ex. 10.0.0.0 to 10.2.0.0) ....................... 7 Licensing the desktop client ............................................................................................................. 9 Upgrading the Database ................................................................................................................ 10 Database Server Upgrade ........................................................................................................................... 11 Upgrading the Database Server ..................................................................................................... 11 Changing the Database Service ...................................................................................................... 14 Web Server Upgrade .................................................................................................................................. -

Download Issue
BRAZIL: Booming, Busting … and Now? TALENT + LEADERSHIP ISSUE NO. 3 0 Are Leaders Doomed? The Coolest Jobs $14.95 US / CAN / $14.95 US Will Humans Matter? IT’S WHERE YOUR GREATEST SOURCE OF VALUE CAN DRIVE YOUR BUSINESS PERFORMANCE. Maximizing performance in your organization today and tomorrow comes down to releasing the full potential of your greatest source of value. And, contrary to what many may believe, technology isn’t it. At Korn Ferry, we conducted in depth, quantifiable research to uncover the truth about the future of work. Are you placing your bets wisely? Get your organization to UP. Kornferry.com/futureofwork IT’S WHERE YOUR GREATEST SOURCE OF VALUE CAN DRIVE YOUR BUSINESS PERFORMANCE. Maximizing performance in your organization today and tomorrow comes down to releasing the full potential of your greatest source of value. And, contrary to what many may believe, technology isn’t it. At Korn Ferry, we conducted in depth, quantifiable research to uncover the truth about the future of work. Are you placing your bets wisely? Get your organization to UP. Kornferry.com/futureofwork Gary Burnison Thought leadership. Timely insights. And more. Chief Executive Officer UP. IT’S WHERE STRATEGY AND kornferryinstitute.com Michael Distefano PURPOSE MEET PEOPLE Chief Marketing Officer & President, Korn Ferry Institute AND EXECUTION. The Toughest Job to Fill Jonathan Dahl The improving economy has made qualified Editor-in-Chief You’re looking for new pathways to drive talent harder to find. You’ll never guess the sustainable, profi table growth. role that thousands of the world’s top talent Russell Pearlman acquisition professionals say is now the toughest Managing Editor to recruit. -

Open Source Used in Cisco Prime Access Registrar 8.0
Open Source Used In Cisco Prime Access Registrar 8.0 Cisco Systems, Inc. www.cisco.com Cisco has more than 200 offices worldwide. Addresses, phone numbers, and fax numbers are listed on the Cisco website at www.cisco.com/go/offices. Text Part Number: 78EE117C99-161538199 Open Source Used In Cisco Prime Access Registrar 8.0 1 This document contains licenses and notices for open source software used in this product. With respect to the free/open source software listed in this document, if you have any questions or wish to receive a copy of any source code to which you may be entitled under the applicable free/open source license(s) (such as the GNU Lesser/General Public License), please contact us at [email protected]. In your requests please include the following reference number 78EE117C99-161538199 Contents 1.1 ace-5.5 5.5 1.1.1 Available under license 1.2 ajaxanywhere 1.2.1 1.2.1 Available under license 1.3 ant 1.7.1 1.3.1 Available under license 1.4 Apache Commons Logging 1.1.1 1.4.1 Available under license 1.5 Apache Tomcat 8.5.16 1.5.1 Available under license 1.6 axis 1.4 1.6.1 Available under license 1.7 Axis C++ 1.6b 1.7.1 Available under license 1.8 Bean Validation API 1.1.0.Final 1.8.1 Available under license 1.9 c-ares 1.10.0 1.9.1 Available under license 1.10 CGLIB 2.2 1.10.1 Available under license 1.11 cglib 2.2.0-b21 :repackaged as a module 1.11.1 Available under license 1.12 cglib 2.1.88 1.12.1 Available under license 1.13 ciscosafec-3.0.0 3.0.0 1.13.1 Available under license Open Source Used In Cisco -

Licensing Information User Manual Mysql Enterprise Monitor 3.4
Licensing Information User Manual MySQL Enterprise Monitor 3.4 Table of Contents Licensing Information .......................................................................................................................... 4 Licenses for Third-Party Components .................................................................................................. 5 Ant-Contrib ............................................................................................................................... 10 ANTLR 2 .................................................................................................................................. 11 ANTLR 3 .................................................................................................................................. 11 Apache Commons BeanUtils v1.6 ............................................................................................. 12 Apache Commons BeanUtils v1.7.0 and Later ........................................................................... 13 Apache Commons Chain .......................................................................................................... 13 Apache Commons Codec ......................................................................................................... 13 Apache Commons Collections .................................................................................................. 14 Apache Commons Daemon ...................................................................................................... 14 Apache -

Licensed to the Apache Software Foundation (ASF) Under One Or More Contributor License Agreements
================================================================================ Licensed to the Apache Software Foundation (ASF) under one or more contributor license agreements. See the NOTICE file distributed with this work for additional information regarding copyright ownership. The ASF licenses this file to You under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at http://www.apache.org/licenses/LICENSE-2.0 Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License. ================================================================================ =================================================== Running The Apache Tomcat 8.0 Servlet/JSP Container =================================================== Apache Tomcat 8.0 requires a Java Standard Edition Runtime Environment (JRE) version 7 or later. ============================= Running With JRE 7 Or Later ============================= (1) Download and Install a Java SE Runtime Environment (JRE) (1.1) Download a Java SE Runtime Environment (JRE), release version 7 or later, from http://www.oracle.com/technetwork/java/javase/downloads/index.html (1.2) Install the JRE according to the instructions included with the release. You may also use a full Java Development Kit (JDK) rather than just a JRE. (2) Download and Install Apache Tomcat (2.1) Download a binary distribution of Tomcat from: http://tomcat.apache.org/ (2.2) Unpack the binary distribution so that it resides in its own directory (conventionally named "apache-tomcat-[version]"). For the purposes of the remainder of this document, the name "CATALINA_HOME" is used to refer to the full pathname of that directory. -
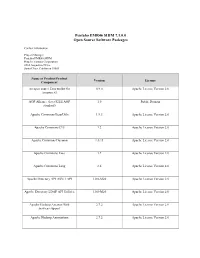
Pentaho EMR46 SHIM 7.1.0.0 Open Source Software Packages
Pentaho EMR46 SHIM 7.1.0.0 Open Source Software Packages Contact Information: Project Manager Pentaho EMR46 SHIM Hitachi Vantara Corporation 2535 Augustine Drive Santa Clara, California 95054 Name of Product/Product Version License Component An open source Java toolkit for 0.9.0 Apache License Version 2.0 Amazon S3 AOP Alliance (Java/J2EE AOP 1.0 Public Domain standard) Apache Commons BeanUtils 1.9.3 Apache License Version 2.0 Apache Commons CLI 1.2 Apache License Version 2.0 Apache Commons Daemon 1.0.13 Apache License Version 2.0 Apache Commons Exec 1.2 Apache License Version 2.0 Apache Commons Lang 2.6 Apache License Version 2.0 Apache Directory API ASN.1 API 1.0.0-M20 Apache License Version 2.0 Apache Directory LDAP API Utilities 1.0.0-M20 Apache License Version 2.0 Apache Hadoop Amazon Web 2.7.2 Apache License Version 2.0 Services support Apache Hadoop Annotations 2.7.2 Apache License Version 2.0 Name of Product/Product Version License Component Apache Hadoop Auth 2.7.2 Apache License Version 2.0 Apache Hadoop Common - 2.7.2 Apache License Version 2.0 org.apache.hadoop:hadoop-common Apache Hadoop HDFS 2.7.2 Apache License Version 2.0 Apache HBase - Client 1.2.0 Apache License Version 2.0 Apache HBase - Common 1.2.0 Apache License Version 2.0 Apache HBase - Hadoop 1.2.0 Apache License Version 2.0 Compatibility Apache HBase - Protocol 1.2.0 Apache License Version 2.0 Apache HBase - Server 1.2.0 Apache License Version 2.0 Apache HBase - Thrift - 1.2.0 Apache License Version 2.0 org.apache.hbase:hbase-thrift Apache HttpComponents Core -
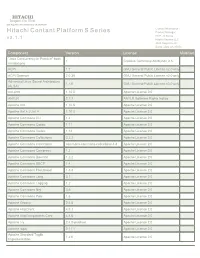
Hitachi Contant Platform S Series Product Manager HCP - S Series V 3
HITACHI Inspire the Next 2535 Augustine Drive Santa Clara, CA 95054 USA Contact Information : Hitachi Contant Platform S Series Product Manager HCP - S Series v 3 . 1 . 1 Hitachi Vantara LLC 2535 Augustine Dr. Santa Clara CA 95054 Component Version License Modified "Java Concurrency in Practice" book 1 Creative Commons Attribution 2.5 annotations ACPI 1.7 GNU General Public License v2.0 only ACPI Daemon 2.0.30 GNU General Public License v2.0 only Advanced Linux Sound Architecture 1.1.6 GNU General Public License v3.0 only (ALSA) ant-antlr 1.10.5 Apache License 2.0 ANTLR 2.7.7 ANTLR Software Rights Notice Apache Ant 1.10.5 Apache License 2.0 Apache Ant + JUnit 4 1.10.5 Apache License 2.0 Apache Commons CLI 1.4 Apache License 2.0 Apache Commons Codec 1.11 Apache License 2.0 Apache Commons Codec 1.14 Apache License 2.0 Apache Commons Collections 3.2.2 Apache License 2.0 Apache Commons Collections commons-commons-collections-4.4 Apache License 2.0 Apache Commons Compress 1.2 Apache License 2.0 Apache Commons Daemon 1.2.2 Apache License 2.0 Apache Commons DBCP 1.4 Apache License 2.0 Apache Commons FileUpload 1.3.3 Apache License 2.0 Apache Commons Lang 3.1 Apache License 2.0 Apache Commons Logging 1.2 Apache License 2.0 Apache Commons Net 3.6 Apache License 2.0 Apache Commons Pool 1.6 Apache License 2.0 Apache Groovy 2.4.8 Apache License 2.0 Apache HttpClient 4.5.3 Apache License 2.0 Apache HttpComponents Core 4.4.6 Apache License 2.0 Apache Ivy 2.4.0-patched Apache License 2.0 Apache log4j 2.11.1 Apache License 2.0 Apache Standard Taglib -

Eyeglass Search OSS Licenses and Packages V9
ECA 15.1 opensuse https://en.opensuse.org/openSUSE:License Package Licence Name Version Type Key Licence Name opensuse 15.1 OS annogen:annogen 0.1.0 JAR Not Found antlr:antlr 2.7.7 JAR BSD Berkeley Software Distribution (BSD) aopalliance:aopalliance 1 JAR Public DomainPublic Domain asm:asm 3.1 JAR Not Found axis:axis 1.4 JAR Apache-2.0 The Apache Software License, Version 2.0 axis:axis-wsdl4j 1.5.1 JAR Not Found backport-util-concurrent:backport-util-concurrent3.1 JAR Public DomainPublic Domain com.amazonaws:aws-java-sdk 1.1.7.1 JAR Apache-2.0 The Apache Software License, Version 2.0 com.beust:jcommander 1.72 JAR Apache-2.0 The Apache Software License, Version 2.0 com.carrotsearch:hppc 0.6.0 JAR Apache-2.0 The Apache Software License, Version 2.0 com.clearspring.analytics:stream 2.7.0 JAR Apache-2.0 The Apache Software License, Version 2.0 com.drewnoakes:metadata-extractor 2.4.0-beta-1JAR Public DomainPublic Domain com.esotericsoftware:kryo-shaded 3.0.3 JAR BSD Berkeley Software Distribution (BSD) com.esotericsoftware:minlog 1.3.0 JAR BSD Berkeley Software Distribution (BSD) com.fasterxml.jackson.core:jackson-annotations2.6.1 JAR Apache-2.0 The Apache Software License, Version 2.0 com.fasterxml.jackson.core:jackson-annotations2.5.0 JAR Apache-2.0 The Apache Software License, Version 2.0 com.fasterxml.jackson.core:jackson-annotations2.2.0 JAR Apache-2.0 The Apache Software License, Version 2.0 com.fasterxml.jackson.core:jackson-annotations2.9.0 JAR Apache-2.0 The Apache Software License, Version 2.0 com.fasterxml.jackson.core:jackson-annotations2.6.0 -
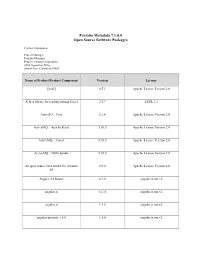
Pentaho Metadata 7.1.0.0 Open Source Software Packages
Pentaho Metadata 7.1.0.0 Open Source Software Packages Contact Information: Project Manager Pentaho Metadata Hitachi Vantara Corporation 2535 Augustine Drive Santa Clara, California 95054 Name of Product/Product Component Version License [ini4j] 0.5.1 Apache License Version 2.0 A Java library for reading/writing Excel 2.5.7 LGPL 2.1 ActiveIO :: Core 3.1.4 Apache License Version 2.0 ActiveMQ :: Apache Karaf 5.10.0 Apache License Version 2.0 ActiveMQ :: Camel 5.10.0 Apache License Version 2.0 ActiveMQ :: OSGi bundle 5.10.0 Apache License Version 2.0 An open source Java toolkit for Amazon 0.9.0 Apache License Version 2.0 S3 Angular UI Router 0.3.0 angular.js.mit.v2 angular.js 1.2.15 angular.js.mit.v2 angular.js 1.5.8 angular.js.mit.v2 angular-animate-1.5.6 1.5.6 angular.js.mit.v2 Name of Product/Product Component Version License angular-route 1.5.6 angular.js.mit.v2 angular-sanitize-1.5.6 1.5.6 angular.js.mit.v2 angular-translate 2.3.0 angular.js.mit.v2 angular-translate-2.12.1 2.12.1 angular.bootstrap.mit.v2 angular-translate-2.12.1 2.12.1 angular.js.mit.v2 AngularUI Bootstrap 0.6.0 angular.bootstrap.mit.v2 AngularUI Bootstrap 1.3.3 angular.bootstrap.mit.v2 Annotation 1.0 1.1.1 Apache License Version 2.0 Annotation 1.1 1.0.1 Apache License Version 2.0 ANTLR 3 Complete 3.5.2 ANTLR License Antlr 3.4 Runtime 3.4 ANTLR License ANTLR, ANother Tool for Language 2.7.7 ANTLR License Recognition AOP Alliance (Java/J2EE AOP standard) 1.0 Public Domain Apache Ant Core 1.9.1 Apache License Version 2.0 Name of Product/Product Component Version License