Analysis of Y-Chromosome Polymorphisms in Pakistani
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Germanic Origins from the Perspective of the Y-Chromosome
Germanic Origins from the Perspective of the Y-Chromosome By Michael Robert St. Clair A dissertation submitted in partial satisfaction of the requirements for the degree of Doctor in Philosophy in German in the Graduate Division of the University of California, Berkeley Committee in charge: Irmengard Rauch, Chair Thomas F. Shannon Montgomery Slatkin Spring 2012 Abstract Germanic Origins from the Perspective of the Y-Chromosome by Michael Robert St. Clair Doctor of Philosophy in German University of California, Berkeley Irmengard Rauch, Chair This dissertation holds that genetic data are a useful tool for evaluating contemporary models of Germanic origins. The Germanic languages are a branch of the Indo-European language family and include among their major contemporary representatives English, German, Dutch, Danish, Swedish, Norwegian and Icelandic. Historically, the search for Germanic origins has sought to determine where the Germanic languages evolved, and why the Germanic languages are similar to and different from other European languages. Both archaeological and linguist approaches have been employed in this research direction. The linguistic approach to Germanic origins is split among those who favor the Stammbaum theory and those favoring language contact theory. Stammbaum theory posits that Proto-Germanic separated from an ancestral Indo-European parent language. This theoretical approach accounts for similarities between Germanic and other Indo- European languages by posting a period of mutual development. Germanic innovations, on the other hand, occurred in isolation after separation from the parent language. Language contact theory posits that Proto-Germanic was the product of language convergence and this convergence explains features that Germanic shares with other Indo-European languages. -

University of Huddersfield Repository
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by University of Huddersfield Repository University of Huddersfield Repository Fornarino, Simona, Pala, Maria, Battaglia, Vincenza, Maranta, Ramona, Achilli, Alessandro, Modiano, Guido, Torroni, Antonio, Semino, Ornella and Santachiara-Benerecetti, Silvana A Mitochondrial and Y-chromosome diversity of the Tharus (Nepal): a reservoir of genetic variation Original Citation Fornarino, Simona, Pala, Maria, Battaglia, Vincenza, Maranta, Ramona, Achilli, Alessandro, Modiano, Guido, Torroni, Antonio, Semino, Ornella and Santachiara-Benerecetti, Silvana A (2009) Mitochondrial and Y-chromosome diversity of the Tharus (Nepal): a reservoir of genetic variation. BMC Evolutionary Biology, 9 (1). p. 154. ISSN 1471-2148 This version is available at http://eprints.hud.ac.uk/15482/ The University Repository is a digital collection of the research output of the University, available on Open Access. Copyright and Moral Rights for the items on this site are retained by the individual author and/or other copyright owners. Users may access full items free of charge; copies of full text items generally can be reproduced, displayed or performed and given to third parties in any format or medium for personal research or study, educational or not-for-profit purposes without prior permission or charge, provided: • The authors, title and full bibliographic details is credited in any copy; • A hyperlink and/or URL is included for the original metadata page; and • The content is -
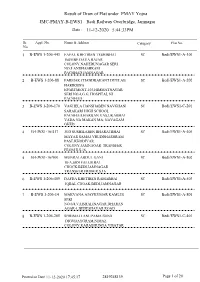
Jmc-Pmay-B-Ews1 11-12-2020 5:44:33Pm
Result of Draw of Flat under PMAY Yojna JMC-PMAY-B-EWS1 Bedi Railway Overbridge, Jamnagar Date : 11-12-2020 5:44:33PM Sr. Appl. No. Name & Address Category Flat No. No. 1 B-EWS 1-206-443 FAFAL KHETIBEN TESHIBHAI SC Bedi/EWS1-A-106 BOMBE DAVA BAJAR COLONY,NAHERUNAGAR SERI NO.5 ANDHASHRAM PACHHAD,JAMNAGAR 2 B-EWS 1-206-88 PARMAR CHANDRAKANT DHULAB SC Bedi/EWS1-A-202 HARIKRIPA EPARTMENT-103,HIMMATNAGAR SERI NO.4,G.G. HOSPITAL NI PACHHAD 3 B-EWS 1-206-170 VAGHELA HANSHABEN NAVGHANB SC Bedi/EWS1-C-201 SARAKARI HIGH SCHOOL PACHHAD,HARIJAN VAS,UKABHAI VADA NA MAKAN MA, NAVAGAM GHED 4 B-EWS1-16/417 JOD SUSHILABEN BHARATBHAI SC Bedi/EWS1-A-406 MAYAR SAMAJ,VRUDDHASHRAM PASE,KHODIYAR COLONY,JAMNAGAR TRANSFER FROM R.S.16 5 B-EWS1-16/900 MUNRAI ABDUL GANI SC Bedi/EWS1-A-802 18-A,SIDI FALI,IKBAL CHOCK,BEDI,JAMNAGAR TRANSFER FROM R.S.16 6 B-EWS 1-206-409 DAFDA KHETIBEN RAMABHAI SC Bedi/EWS1-A-503 IQBAL CHOAK,BEDI,JAMNAGAR 7 B-EWS 1-206-14 MAKVANA AJAYKUMAR KAMLES SC Bedi/EWS1-A-801 SERI NO.6/4,VAISHALINAGAR,DHARAN AGAR-1,BEDESHAVAR ROAD 8 B-EWS 1-206-205 SHRIMALI ANUPAMA SUNIL SC Bedi/EWS1-C-401 DIGVIJAYGRAM,NIMAZ COLONY,KARABHUNGA VISATAR Printed on Date 11-12-2020 17:45:17 2819248139 Page 1 of 20 Result of Draw of Flat under PMAY Yojna JMC-PMAY-B-EWS1 Bedi Railway Overbridge, Jamnagar Date : 11-12-2020 5:44:33PM Sr. -

BMC Genetics Biomed Central
BMC Genetics BioMed Central Research article Open Access Saudi Arabian Y-Chromosome diversity and its relationship with nearby regions Khaled K Abu-Amero*1, Ali Hellani2, Ana M González3, Jose M Larruga3, Vicente M Cabrera3 and Peter A Underhill4 Address: 1Molecular Genetics Laboratory, College of Medicine, King Saud University, Riyadh 11411, Saudi Arabia, 2Department of PGD, Saad Specialist Hospital, Al-Khobar, Saudi Arabia, 3Departamento de Genética, Universidad de La Laguna, 38271 La Laguna, Tenerife, Spain and 4Department of Psychiatry and Behavioural Sciences, Stanford University, School of Medicine, Stanford, California 94304, USA Email: Khaled K Abu-Amero* - [email protected]; Ali Hellani - [email protected]; Ana M González - [email protected]; Jose M Larruga - [email protected]; Vicente M Cabrera - [email protected]; Peter A Underhill - [email protected] * Corresponding author Published: 22 September 2009 Received: 9 December 2008 Accepted: 22 September 2009 BMC Genetics 2009, 10:59 doi:10.1186/1471-2156-10-59 This article is available from: http://www.biomedcentral.com/1471-2156/10/59 © 2009 Abu-Amero et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: Human origins and migration models proposing the Horn of Africa as a prehistoric exit route to Asia have stimulated molecular genetic studies in the region using uniparental loci. However, from a Y-chromosome perspective, Saudi Arabia, the largest country of the region, has not yet been surveyed. -

Custodians of Culture and Biodiversity
Custodians of culture and biodiversity Indigenous peoples take charge of their challenges and opportunities Anita Kelles-Viitanen for IFAD Funded by the IFAD Innovation Mainstreaming Initiative and the Government of Finland The opinions expressed in this manual are those of the authors and do not nec - essarily represent those of IFAD. The designations employed and the presenta - tion of material in this publication do not imply the expression of any opinion whatsoever on the part of IFAD concerning the legal status of any country, terri - tory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. The designations “developed” and “developing” countries are in - tended for statistical convenience and do not necessarily express a judgement about the stage reached in the development process by a particular country or area. This manual contains draft material that has not been subject to formal re - view. It is circulated for review and to stimulate discussion and critical comment. The text has not been edited. On the cover, a detail from a Chinese painting from collections of Anita Kelles-Viitanen CUSTODIANS OF CULTURE AND BIODIVERSITY Indigenous peoples take charge of their challenges and opportunities Anita Kelles-Viitanen For IFAD Funded by the IFAD Innovation Mainstreaming Initiative and the Government of Finland Table of Contents Executive summary 1 I Objective of the study 2 II Results with recommendations 2 1. Introduction 2 2. Poverty 3 3. Livelihoods 3 4. Global warming 4 5. Land 5 6. Biodiversity and natural resource management 6 7. Indigenous Culture 7 8. Gender 8 9. -

Genetic Analysis of the Major Tribes of Buner and Swabi Areas Through Dental Morphology and Dna Analysis
GENETIC ANALYSIS OF THE MAJOR TRIBES OF BUNER AND SWABI AREAS THROUGH DENTAL MORPHOLOGY AND DNA ANALYSIS MUHAMMAD TARIQ DEPARTMENT OF GENETICS HAZARA UNIVERSITY MANSEHRA 2017 I HAZARA UNIVERSITY MANSEHRA Department of Genetics GENETIC ANALYSIS OF THE MAJOR TRIBES OF BUNER AND SWABI AREAS THROUGH DENTAL MORPHOLOGY AND DNA ANALYSIS By Muhammad Tariq This research study has been conducted and reported as partial fulfillment of the requirements of PhD degree in Genetics awarded by Hazara University Mansehra, Pakistan Mansehra The Friday 17, February 2017 I ABSTRACT This dissertation is part of the Higher Education Commission of Pakistan (HEC) funded project, “Enthnogenetic elaboration of KP through Dental Morphology and DNA analysis”. This study focused on five major ethnic groups (Gujars, Jadoons, Syeds, Tanolis, and Yousafzais) of Buner and Swabi Districts, Khyber Pakhtunkhwa Province, Pakistan, through investigations of variations in morphological traits of the permanent tooth crown, and by molecular anthropology based on mitochondrial and Y-chromosome DNA analyses. The frequencies of seven dental traits, of the Arizona State University Dental Anthropology System (ASUDAS) were scored as 17 tooth- trait combinations for each sample, encompassing a total sample size of 688 individuals. These data were compared to data collected in an identical fashion among samples of prehistoric inhabitants of the Indus Valley, southern Central Asia, and west-central peninsular India, as well as to samples of living members of ethnic groups from Abbottabad, Chitral, Haripur, and Mansehra Districts, Khyber Pakhtunkhwa and to samples of living members of ethnic groups residing in Gilgit-Baltistan. Similarities in dental trait frequencies were assessed with C.A.B. -

Ethnographic Series, Sidhi, Part IV-B, No-1, Vol-V
CENSUS OF INDIA 1961 VOLUMEV, PART IV-B, No.1 ETHNOGRAPHIC SERIES GUJARAT Preliminary R. M. V ANKANI, investigation Tabulation Officer, and draft: Office of the CensuS Superintendent, Gujarat. SID I Supplementary V. A. DHAGIA, A NEGROID L IBE investigation: Tabulation Officer, Office of the Census Superintendent, OF GU ARAT Gujarat. M. L. SAH, Jr. Investigator, Office of the Registrar General, India. Fieta guidance, N. G. NAG, supervision and Research Officer, revised draft: Office of the Registrar General, India. Editors: R. K. TRIVEDI, Su perintendent of Census Operations, Gujarat. B. K. Roy BURMAN, Officer on Special Duty, (Handicrafts and Social Studies), Office of the Registrar General, India. K. F. PATEL, R. K. TRIVEDI Deputy Superintendent of Census Superintendent of Census Operations, Gujarat. Operations, Gujarat N. G. NAG, Research Officer, Office' of the Registrar General, India. CENSUS OF INDIA 1961 LIST OF PUBLICATIONS CENTRAL GOVERNMENT PUBLICATIONS Census of India, 1961 Volume V-Gujarat is being published in the following parts: '" I-A(i) General Report '" I-A(ii)a " '" I-A(ii)b " '" I-A(iii) General Report-Economic Trends and Projections :« I-B Report on Vital Statistics and Fertility Survey :I' I-C Subsidiary Tables '" II-A General Population Tables '" II-B(I) General Economic Tables (Tables B-1 to B-IV-C) '" II-B(2) General Economic Tables (Tables B-V to B-IX) '" II-C Cultural and Migration Tables :t< III Household Economic Tables (Tables B-X to B-XVII) "'IV-A Report on Housing and Establishments :t<IV-B Housing and Establishment -

Introduction Since Time Immemorial, Human Beings Have Used Narrative
Chapter 1 – Introduction Since time immemorial, human beings have used narrative to help us make sense of our experience of life. From the fireside to the theatre, from the television and silver screen to the more recent manifestations of the virtual world, we have used storytelling as a means of providing structure, order, and coherence to what can otherwise appear an overwhelming infinity of random, unrelated events. In ordering the perceived chaos of the world around us into a structure we can grasp, narrative provides insight and understanding not only of events themselves, but on a more fundamental level, of the very essence of what it means to live as a human being. As the primary means by which historical writing is organized, narrative has attracted a large body of historians and philosophers who have grappled with its impact on our understanding of the past. Underlying their work is the tension between historical writing as a reflection of what took place in the past, and the essence of narrative as a creative, imaginative act. The very structure of Aristotelian narrative, with its causal link between events, its clearly defined beginning, middle and end, its promise of catharsis, its theme or moral, reflects an act of imagination on the part of its author. While an effective narrative first and foremost strives to draw us into its world of story and keep us there until the ending, the primary goal of historical writing, in theory at least, is to increase our understanding about the past. While these two goals are not inherently incompatible, they do not always work in concert. -

2 О Сен 2012 Москва-2012 Работа Выполнена В Федеральном Государственном Бюджетном Учреждении «Медико-Генетический Научный Це)Гтр» Российской Академии Медицинских Наук
На правах рукописи 005047иоо БАЛАНОВСКИЙ Олег Павлович ИЗМЕНЧИВОСТЬ ГЕНОФОНДА В ПРОСТРАНСТВЕ И ВРЕМЕНИ: СИНТЕЗ ДАННЫХ О ГЕНОГЕОГРАФИИ МИТОХОНДРИАЛЬНОЙ ДНК И У-ХРОМОСОМЫ 03.02.07 - генетика 03.01.03 - молекулярная биология АВТОРЕФЕРАТ диссертации на соискание ученой степени доктора биологических наук 2 О СЕН 2012 Москва-2012 Работа выполнена в Федеральном государственном бюджетном учреждении «Медико-генетический научный це)гтр» Российской академии медицинских наук. Научные консультанты: доктор биологических наук, академик Эстонской АН Виллемс Рихард Лео-Энделевнч; доктор биолошческих наук, член-корреспондент РАН Янковосии Николай Казнмнровнч. Официальные оппоненты: Степанов Вадим Анатольевич, доктор биологических наук, профессор Федеральное государственное бюджетное учреждение «Научно-исследовательский инстшуг медицинской генетики» Сибирского отделения Российской академии медицинских наук, заместитель директора по научным вопросам; Сиицын Виктор Алексеевич, доктор биологических наук, профессор Федеральное государственное бюджетное учреждение «Медико-генетический иаутый це1ггр)) Российской академии медицинских наук, завеодтощий лабораторией экологической генетики; Захаров-Гезехус Илья Артемьевич, доктор биологических наук, профессор, член- корреспондеш-Российской академии наук Федеральное государственное бюджетное учреждение нафтен Институт общей генетики им. Н.И. Вавилова Российской академии наук, советник РАН. Ведущая организация: Федеральное государственное бюджетное образовательное учреждение высшего профессионального образования «Московский -

HUMAN MITOCHONDRIAL DNA HAPLOGROUP J in EUROPE and NEAR EAST M.Sc
UNIVERSITY OF TARTU FACULTY OF BIOLOGY AND GEOGRAPHY, INSTITUTE OF MOLECULAR AND CELL BIOLOGY, DEPARTMENT OF EVOLUTIONARY BIOLOGY Piia Serk HUMAN MITOCHONDRIAL DNA HAPLOGROUP J IN EUROPE AND NEAR EAST M.Sc. Thesis Supervisors: Ph.D. Ene Metspalu, Prof. Richard Villems Tartu 2004 Table of contents Abbreviations .............................................................................................................................3 Definition of basic terms used in the thesis.........................................................................3 Introduction................................................................................................................................4 Literature overview ....................................................................................................................5 West–Eurasian mtDNA tree................................................................................................5 Fast mutation rate of mtDNA..............................................................................................9 Estimation of a coalescence time ......................................................................................10 Topology of mtDNA haplogroup J....................................................................................12 Geographic spread of mtDNA haplogroup J.....................................................................20 The aim of the present study ....................................................................................................22 -

Ancient Mitochondrial DNA from Pre-Historic
Grand Valley State University ScholarWorks@GVSU Masters Theses Graduate Research and Creative Practice 4-30-2011 Ancient Mitochondrial DNA From Pre-historic Southeastern Europe: The rP esence of East Eurasian Haplogroups Provides Evidence of Interactions with South Siberians Across the Central Asian Steppe Belt Jeremy R. Newton Grand Valley State University Follow this and additional works at: http://scholarworks.gvsu.edu/theses Part of the Cell Biology Commons, and the Molecular Biology Commons Recommended Citation Newton, Jeremy R., "Ancient Mitochondrial DNA From Pre-historic Southeastern Europe: The rP esence of East Eurasian Haplogroups Provides Evidence of Interactions with South Siberians Across the Central Asian Steppe Belt" (2011). Masters Theses. 5. http://scholarworks.gvsu.edu/theses/5 This Thesis is brought to you for free and open access by the Graduate Research and Creative Practice at ScholarWorks@GVSU. It has been accepted for inclusion in Masters Theses by an authorized administrator of ScholarWorks@GVSU. For more information, please contact [email protected]. ANCIENT MITOCHONDRIAL DNA FROM PRE-HISTORIC SOUTH- EASTERN EUROPE: THE PRESENCE OF EAST EURASIAN HAPLOGROUPS PROVIDES EVIDENCE OF INTERACTIONS WITH SOUTH SIBERIANS ACROSS THE CENTRAL ASIAN STEPPE BELT A thesis submittal in partial fulfillment of the requirements for the degree of Master of Science By Jeremy R. Newton To Cell and Molecular Biology Department Grand Valley State University Allendale, MI April, 2011 “Not all those who wander are lost.” J.R.R. Tolkien iii ACKNOWLEDGEMENTS I would like to extend my sincerest thanks to every person who has motivated, directed, and encouraged me throughout this thesis project. I especially thank my graduate advisor, Dr. -

Are the Kalasha Really of Greek Origin? the Legend of Alexander the Great and the Pre-Islamic World of the Hindu Kush1
Acta Orientalia 2011: 72, 47–92. Copyright © 2011 Printed in India – all rights reserved ACTA ORIENTALIA ISSN 0001-6483 Are the Kalasha really of Greek origin? The Legend of Alexander the Great and the Pre-Islamic World of the Hindu Kush1 Augusto S. Cacopardo Università di Firenze Abstract The paper refutes the claim that the Kalasha may be the descendants of the Greeks of Asia. First, traditions of Alexandrian descent in the Hindu Kush are examined on the basis of written sources and it is shown that such legends are not part of Kalasha traditional knowledge. Secondly, it is argued that the Kalasha were an integral part of the pre-Islamic cultural fabric of the Hindu Kush, and cannot be seen as intruders in the area, as legends of a Greek descent would want them. Finally, through comparative suggestions, it is proposed that possible similarities between the Kalasha and pre-Christian 1 Paper presented as a key-note address at the First International Conference on Language Documentation and Tradition, with a Special Interest in the Kalasha of the Hindu Kush Valleys, Himalayas – Thessaloniki, Greece, 7–9 November 2008. Scarcity of funds caused the scientific committee to decide to select for the forthcoming proceedings only linguistic papers. This is rather unfortunate because the inclusion in the volume of anthropological papers as well would have offered a good opportunity for comparing different views on the question of the Greek ascendancy of the Kalasha. 48 Augusto S. Cacopardo Europe are to be explained by the common Indo-European heritage rather than by more recent migrations and contacts.