Relevance of Copy Number Variation at Chromosome X in Male Fetuses
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Cancer Immunity (1 December 2006) Vol
Cancer Immun 1424-9634Academy of Cancer Immunology Cancer Immunity (1 December 2006) Vol. 6, p. 12 Submitted: 26 September 2006. Accepted: 10 October 2006. Copyright © 2006 by Andrew J. G. Simpson 061012 Article Physical interaction of two cancer-testis antigens, MAGE-C1 (CT7) and NY-ESO-1 (CT6) Hearn J. Cho1*,**, Otavia L. Caballero2*, Sacha Gnjatic2, Valéria C. C. Andrade3, Gisele W. Colleoni3, Andre L. Vettore4, Hasina H. Outtz1, Sheila Fortunato2, Nasser Altorki1, Cathy A. Ferrera1, Ramon Chua2, Achim A. Jungbluth2, Yao-Tseng Chen1, Lloyd J. Old2 and Andrew J. G. Simpson2 1Weill Medical College of Cornell University, 1300 York Avenue, New York, NY 10021, USA 2Ludwig Institute for Cancer Research, New York Branch at Memorial Sloan-Kettering Cancer Center, 1275 York Avenue, New York, NY 10021, USA 3Escola Paulista de Medicina, Universidade Federal de Sao Paulo, Sao Paulo, SP, Brazil 4Ludwig Institute for Cancer Research, Sao Paulo Branch, Sao Paulo, SP, Brazil *These authors contributed equally to this work **Present address: NYU Cancer Institute, New York University School of Medicine, 550 First Avenue, New York, NY 10016, USA Contributed by: LJ Old Cancer/testis (CT) antigens are the protein products of germ line- encoded on the X chromosome (CT-X antigens) and those that associated genes that are activated in a wide variety of tumors and can are not (non-X CT antigens) (1). elicit autologous cellular and humoral immune responses. CT It is estimated that 10% of the genes on the X-chromosome antigens can be divided between those that are encoded on the X belong to CT-X families (5). -

The HECT Domain Ubiquitin Ligase HUWE1 Targets Unassembled Soluble Proteins for Degradation
OPEN Citation: Cell Discovery (2016) 2, 16040; doi:10.1038/celldisc.2016.40 ARTICLE www.nature.com/celldisc The HECT domain ubiquitin ligase HUWE1 targets unassembled soluble proteins for degradation Yue Xu1, D Eric Anderson2, Yihong Ye1 1Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD, USA; 2Advanced Mass Spectrometry Core Facility, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD, USA In eukaryotes, many proteins function in multi-subunit complexes that require proper assembly. To maintain complex stoichiometry, cells use the endoplasmic reticulum-associated degradation system to degrade unassembled membrane subunits, but how unassembled soluble proteins are eliminated is undefined. Here we show that degradation of unassembled soluble proteins (referred to as unassembled soluble protein degradation, USPD) requires the ubiquitin selective chaperone p97, its co-factor nuclear protein localization protein 4 (Npl4), and the proteasome. At the ubiquitin ligase level, the previously identified protein quality control ligase UBR1 (ubiquitin protein ligase E3 component n-recognin 1) and the related enzymes only process a subset of unassembled soluble proteins. We identify the homologous to the E6-AP carboxyl terminus (homologous to the E6-AP carboxyl terminus) domain-containing protein HUWE1 as a ubiquitin ligase for substrates bearing unshielded, hydrophobic segments. We used a stable isotope labeling with amino acids-based proteomic approach to identify endogenous HUWE1 substrates. Interestingly, many HUWE1 substrates form multi-protein com- plexes that function in the nucleus although HUWE1 itself is cytoplasmically localized. Inhibition of nuclear entry enhances HUWE1-mediated ubiquitination and degradation, suggesting that USPD occurs primarily in the cytoplasm. -

Supplemental Table S1
Entrez Gene Symbol Gene Name Affymetrix EST Glomchip SAGE Stanford Literature HPA confirmed Gene ID Profiling profiling Profiling Profiling array profiling confirmed 1 2 A2M alpha-2-macroglobulin 0 0 0 1 0 2 10347 ABCA7 ATP-binding cassette, sub-family A (ABC1), member 7 1 0 0 0 0 3 10350 ABCA9 ATP-binding cassette, sub-family A (ABC1), member 9 1 0 0 0 0 4 10057 ABCC5 ATP-binding cassette, sub-family C (CFTR/MRP), member 5 1 0 0 0 0 5 10060 ABCC9 ATP-binding cassette, sub-family C (CFTR/MRP), member 9 1 0 0 0 0 6 79575 ABHD8 abhydrolase domain containing 8 1 0 0 0 0 7 51225 ABI3 ABI gene family, member 3 1 0 1 0 0 8 29 ABR active BCR-related gene 1 0 0 0 0 9 25841 ABTB2 ankyrin repeat and BTB (POZ) domain containing 2 1 0 1 0 0 10 30 ACAA1 acetyl-Coenzyme A acyltransferase 1 (peroxisomal 3-oxoacyl-Coenzyme A thiol 0 1 0 0 0 11 43 ACHE acetylcholinesterase (Yt blood group) 1 0 0 0 0 12 58 ACTA1 actin, alpha 1, skeletal muscle 0 1 0 0 0 13 60 ACTB actin, beta 01000 1 14 71 ACTG1 actin, gamma 1 0 1 0 0 0 15 81 ACTN4 actinin, alpha 4 0 0 1 1 1 10700177 16 10096 ACTR3 ARP3 actin-related protein 3 homolog (yeast) 0 1 0 0 0 17 94 ACVRL1 activin A receptor type II-like 1 1 0 1 0 0 18 8038 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 1 0 0 0 0 19 8751 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 1 0 0 0 0 20 8728 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 1 0 0 0 0 21 81792 ADAMTS12 ADAM metallopeptidase with thrombospondin type 1 motif, 12 1 0 0 0 0 22 9507 ADAMTS4 ADAM metallopeptidase with thrombospondin type 1 -
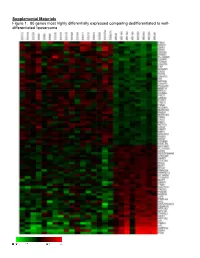
Supplemental Materials Figure 1. 80 Genes Most Highly Differentially Expressed Comparing Dedifferentiated to Well- Differentiated Liposarcoma
Supplemental Materials Figure 1. 80 genes most highly differentially expressed comparing dedifferentiated to well- differentiated liposarcoma Figure 2. Validation of microa CDC2, RACGAP1, FGFR-2, MAD2 and CITED1 200 CDK4 by Liposarcoma Subtype 16 0 12 0 80 Expression40 Level 0 Nl Fat rray data by quantitative RT-P Well Diff 16 0 p16 by Liposarcoma Subtype Dediff 12 0 80 Myxoid Expression40 Level Round Cell 0 12 0 MDM2 by Liposarcoma Subtype 10 0 Pleomorphic Nl Fat 80 60 40 Expression Level 20 RACGAP1 by LiposarcomaWell Diff Subtype 0 70 CR for genes CDK4, MDM2, p16, 60 Dediff 50 Nl Fat 40 30 Myxoid Expression20 Level Well Diff 10 50 0 Round Cell CDC2 by Liposarcoma Subtype Dediff 40 Nl Fat Pleomorphic 30 Myxoid 20 Expression Level Well Diff 10 MAD2L1 by Liposarcoma Subtype Round Cell 60 0 50 Dediff Pleomorphic 40 Nl Fat 30 Myxoid 20 Expression Level Well Diff 10 FGFR-2 by Liposarcoma Subtype 0 Round Cell 200 16 0 Dediff Nl Fat Pleomorphic 12 0 Myxoid 80 Expression Level Well Diff 40 Round Cell 0 Dediff Pleomorphic Nl Fat Myxoid Well Diff CITED1 by Liposarcoma Subtype Round Cell 10 0 80 Dediff Pleomorphic 60 40 Myxoid Expression Level 20 0 Round Cell Nl Fat Pleomorphic Well Diff Dediff Myxoid Round Cell Pleomorphic Table 1. 142 Gene classifier for liposarcoma subtype The genes used for each pair wise subtype comparison are grouped together. The flag column indicates which genes are unique to each subtype comparison. The values show the mean expression levels (actually the mean of the log expression levels was computed and than transformed back to absolute expression level). -

Environmental Influences on Endothelial Gene Expression
ENDOTHELIAL CELL GENE EXPRESSION John Matthew Jeff Herbert Supervisors: Prof. Roy Bicknell and Dr. Victoria Heath PhD thesis University of Birmingham August 2012 University of Birmingham Research Archive e-theses repository This unpublished thesis/dissertation is copyright of the author and/or third parties. The intellectual property rights of the author or third parties in respect of this work are as defined by The Copyright Designs and Patents Act 1988 or as modified by any successor legislation. Any use made of information contained in this thesis/dissertation must be in accordance with that legislation and must be properly acknowledged. Further distribution or reproduction in any format is prohibited without the permission of the copyright holder. ABSTRACT Tumour angiogenesis is a vital process in the pathology of tumour development and metastasis. Targeting markers of tumour endothelium provide a means of targeted destruction of a tumours oxygen and nutrient supply via destruction of tumour vasculature, which in turn ultimately leads to beneficial consequences to patients. Although current anti -angiogenic and vascular targeting strategies help patients, more potently in combination with chemo therapy, there is still a need for more tumour endothelial marker discoveries as current treatments have cardiovascular and other side effects. For the first time, the analyses of in-vivo biotinylation of an embryonic system is performed to obtain putative vascular targets. Also for the first time, deep sequencing is applied to freshly isolated tumour and normal endothelial cells from lung, colon and bladder tissues for the identification of pan-vascular-targets. Integration of the proteomic, deep sequencing, public cDNA libraries and microarrays, delivers 5,892 putative vascular targets to the science community. -

Stelios Pavlidis3, Matthew Loza3, Fred Baribaud3, Anthony
Supplementary Data Th2 and non-Th2 molecular phenotypes of asthma using sputum transcriptomics in UBIOPRED Chih-Hsi Scott Kuo1.2, Stelios Pavlidis3, Matthew Loza3, Fred Baribaud3, Anthony Rowe3, Iaonnis Pandis2, Ana Sousa4, Julie Corfield5, Ratko Djukanovic6, Rene 7 7 8 2 1† Lutter , Peter J. Sterk , Charles Auffray , Yike Guo , Ian M. Adcock & Kian Fan 1†* # Chung on behalf of the U-BIOPRED consortium project team 1Airways Disease, National Heart & Lung Institute, Imperial College London, & Biomedical Research Unit, Biomedical Research Unit, Royal Brompton & Harefield NHS Trust, London, United Kingdom; 2Department of Computing & Data Science Institute, Imperial College London, United Kingdom; 3Janssen Research and Development, High Wycombe, Buckinghamshire, United Kingdom; 4Respiratory Therapeutic Unit, GSK, Stockley Park, United Kingdom; 5AstraZeneca R&D Molndal, Sweden and Areteva R&D, Nottingham, United Kingdom; 6Faculty of Medicine, Southampton University, Southampton, United Kingdom; 7Faculty of Medicine, University of Amsterdam, Amsterdam, Netherlands; 8European Institute for Systems Biology and Medicine, CNRS-ENS-UCBL, Université de Lyon, France. †Contributed equally #Consortium project team members are listed under Supplementary 1 Materials *To whom correspondence should be addressed: [email protected] 2 List of the U-BIOPRED Consortium project team members Uruj Hoda & Christos Rossios, Airways Disease, National Heart & Lung Institute, Imperial College London, UK & Biomedical Research Unit, Biomedical Research Unit, Royal -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Ageing-Associated Changes in DNA Methylation in X and Y Chromosomes
Kananen and Marttila Epigenetics & Chromatin (2021) 14:33 Epigenetics & Chromatin https://doi.org/10.1186/s13072-021-00407-6 RESEARCH Open Access Ageing-associated changes in DNA methylation in X and Y chromosomes Laura Kananen1,2,3,4* and Saara Marttila4,5* Abstract Background: Ageing displays clear sexual dimorphism, evident in both morbidity and mortality. Ageing is also asso- ciated with changes in DNA methylation, but very little focus has been on the sex chromosomes, potential biological contributors to the observed sexual dimorphism. Here, we sought to identify DNA methylation changes associated with ageing in the Y and X chromosomes, by utilizing datasets available in data repositories, comprising in total of 1240 males and 1191 females, aged 14–92 years. Results: In total, we identifed 46 age-associated CpG sites in the male Y, 1327 age-associated CpG sites in the male X, and 325 age-associated CpG sites in the female X. The X chromosomal age-associated CpGs showed signifcant overlap between females and males, with 122 CpGs identifed as age-associated in both sexes. Age-associated X chro- mosomal CpGs in both sexes were enriched in CpG islands and depleted from gene bodies and showed no strong trend towards hypermethylation nor hypomethylation. In contrast, the Y chromosomal age-associated CpGs were enriched in gene bodies, and showed a clear trend towards hypermethylation with age. Conclusions: Signifcant overlap in X chromosomal age-associated CpGs identifed in males and females and their shared features suggest that despite the uneven chromosomal dosage, diferences in ageing-associated DNA methylation changes in the X chromosome are unlikely to be a major contributor of sex dimorphism in ageing. -

Microduplication of Xp22.31 and MECP2 Pathogenic Variant in a Girl with Rett Syndrome: a Case Report
IJMS Vol 44, No 4, July 2019 Case Report Microduplication of Xp22.31 and MECP2 Pathogenic Variant in a Girl with Rett Syndrome: A Case Report Estephania Candelo1, MD, MSc; Abstract Diana Ramirez-Montaño1, MD, MSc; Harry Pachajoa1,2, MD, PhD Rett syndrome (RS) is a neurodevelopmental infantile disease characterized by an early normal psychomotor development followed by a regression in the acquisition of normal developmental stages. In the majority of cases, it leads to a sporadic mutation in the 1Center for Research on Congenital MECP2 gene, which is located on the X chromosome. However, Anomalies and Rare Diseases (CIACER), Health Sciences Faculty, this syndrome has also been associated with microdeletions, gene L Building, Universidad Icesi, Cali, translocations, and other gene mutations. A 12-year-old female Colombia; Colombian patient was presented with refractory epilepsy and 2Department of Genetics, Fundación Valle del Lili, Cali, Colombia regression in skill acquisition (especially language with motor and verbal stereotypies, hyperactivity, and autistic spectrum Correspondence: Harry Pachajoa, MD, PhD; disorder criteria). The patient was born to non-consanguineous Center for Research on Congenital parents and had an early normal development until the age of 36 Anomalies and Rare Diseases months. Comparative genomic hybridization array-CGH (750K) (CIACER), Health Sciences Faculty, L Building, ZIP: 760031, Universidad Icesi, was performed and Xp22.31 duplication was detected (6866889- Cali, Colombia 8115153) with a size of 1.248 Mb associated with developmental Tel: +57 2 5552334, Ext: 8075 delay, epilepsy, and autistic traits. Given the clinical criteria of Fax: +57 2 5551441 Email: [email protected] RS, MECP2 sequencing was performed which showed a de novo Received: 03 June 2018 pathogenic variant c.338C>G (p.Pro113Arg). -

Expression of Cancer-Testis Antigens MAGEA1, MAGEA3, ACRBP, PRAME, SSX2, and CTAG2 in Myxoid and Round Cell Liposarcoma
Modern Pathology (2014) 27, 1238–1245 1238 & 2014 USCAP, Inc All rights reserved 0893-3952/14 $32.00 Expression of cancer-testis antigens MAGEA1, MAGEA3, ACRBP, PRAME, SSX2, and CTAG2 in myxoid and round cell liposarcoma Jessica A Hemminger1, Amanda Ewart Toland2, Thomas J Scharschmidt3, Joel L Mayerson3, Denis C Guttridge2 and O Hans Iwenofu1 1Department of Pathology and Laboratory Medicine, Wexner Medical Center at The Ohio State University, Columbus, OH, USA; 2Department of Molecular Virology, Immunology and Medical Genetics, The Ohio State University Wexner Medical Center, Columbus, OH, USA and 3Department of Orthopedics, The Ohio State University Wexner Medical Center, Columbus, OH, USA Myxoid and round-cell liposarcoma is a frequently encountered liposarcoma subtype. The mainstay of treatment remains surgical excision with or without chemoradiation. However, treatment options are limited in the setting of metastatic disease. Cancer-testis antigens are immunogenic antigens with the expression largely restricted to testicular germ cells and various malignancies, making them attractive targets for cancer immunotherapy. Gene expression studies have reported the expression of various cancer-testis antigens in liposarcoma, with mRNA expression of CTAG1B, CTAG2, MAGEA9, and PRAME described specifically in myxoid and round-cell liposarcoma. Herein, we further explore the expression of the cancer-testis antigens MAGEA1, ACRBP, PRAME, and SSX2 in myxoid and round-cell liposarcoma by immunohistochemistry in addition to determining mRNA levels of CTAG2 (LAGE-1), PRAME, and MAGEA3 by quantitative real-time PCR. Samples in formalin-fixed paraffin-embedded blocks (n ¼ 37) and frozen tissue (n ¼ 8) were obtained for immunohistochemistry and quantitative real-time PCR, respectively. Full sections were stained with antibodies to MAGEA1, ACRBP, PRAME, and SSX2 and staining was assessed for intensity (1–2 þ ) and percent tumor positivity. -

Atrazine and Cell Death Symbol Synonym(S)
Supplementary Table S1: Atrazine and Cell Death Symbol Synonym(s) Entrez Gene Name Location Family AR AIS, Andr, androgen receptor androgen receptor Nucleus ligand- dependent nuclear receptor atrazine 1,3,5-triazine-2,4-diamine Other chemical toxicant beta-estradiol (8R,9S,13S,14S,17S)-13-methyl- Other chemical - 6,7,8,9,11,12,14,15,16,17- endogenous decahydrocyclopenta[a]phenanthrene- mammalian 3,17-diol CGB (includes beta HCG5, CGB3, CGB5, CGB7, chorionic gonadotropin, beta Extracellular other others) CGB8, chorionic gonadotropin polypeptide Space CLEC11A AW457320, C-type lectin domain C-type lectin domain family 11, Extracellular growth factor family 11, member A, STEM CELL member A Space GROWTH FACTOR CYP11A1 CHOLESTEROL SIDE-CHAIN cytochrome P450, family 11, Cytoplasm enzyme CLEAVAGE ENZYME subfamily A, polypeptide 1 CYP19A1 Ar, ArKO, ARO, ARO1, Aromatase cytochrome P450, family 19, Cytoplasm enzyme subfamily A, polypeptide 1 ESR1 AA420328, Alpha estrogen receptor,(α) estrogen receptor 1 Nucleus ligand- dependent nuclear receptor estrogen C18 steroids, oestrogen Other chemical drug estrogen receptor ER, ESR, ESR1/2, esr1/esr2 Nucleus group estrone (8R,9S,13S,14S)-3-hydroxy-13-methyl- Other chemical - 7,8,9,11,12,14,15,16-octahydro-6H- endogenous cyclopenta[a]phenanthren-17-one mammalian G6PD BOS 25472, G28A, G6PD1, G6PDX, glucose-6-phosphate Cytoplasm enzyme Glucose-6-P Dehydrogenase dehydrogenase GATA4 ASD2, GATA binding protein 4, GATA binding protein 4 Nucleus transcription TACHD, TOF, VSD1 regulator GHRHR growth hormone releasing -

Mouse Gm44504 Knockout Project (CRISPR/Cas9)
https://www.alphaknockout.com Mouse Gm44504 Knockout Project (CRISPR/Cas9) Objective: To create a Gm44504 knockout Mouse model (C57BL/6J) by CRISPR/Cas-mediated genome engineering. Strategy summary: The Gm44504 gene (NCBI Reference Sequence: NM_001278271 ; Ensembl: ENSMUSG00000015290 ) is located on Mouse chromosome X. 7 exons are identified, with the ATG start codon in exon 4 and the TAG stop codon in exon 7 (Transcript: ENSMUST00000178691). Exon 6 will be selected as target site. Cas9 and gRNA will be co-injected into fertilized eggs for KO Mouse production. The pups will be genotyped by PCR followed by sequencing analysis. Note: Exon 6 starts from about 32.91% of the coding region. Exon 6 covers 44.37% of the coding region. The size of effective KO region: ~209 bp. The KO region does not have any other known gene. Page 1 of 9 https://www.alphaknockout.com Overview of the Targeting Strategy Wildtype allele 5' gRNA region gRNA region 3' 1 6 7 Legends Exon of mouse Gm44504 Knockout region Page 2 of 9 https://www.alphaknockout.com Overview of the Dot Plot (up) Window size: 15 bp Forward Reverse Complement Sequence 12 Note: The 209 bp section of Exon 6 is aligned with itself to determine if there are tandem repeats. No significant tandem repeat is found in the dot plot matrix. So this region is suitable for PCR screening or sequencing analysis. Overview of the Dot Plot (down) Window size: 15 bp Forward Reverse Complement Sequence 12 Note: The 209 bp section of Exon 6 is aligned with itself to determine if there are tandem repeats.