Herpetological Journal FULL PAPER
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

2010-Geo-033 Environmental Scoping Statement
ENVIRONMENTAL SCOPING STATEMENT: FOR ONI WATER SUPPLY GEORGIA MUNICIPAL INFRASTRUCTURE AND IDP HOUSING REHABILITATION PROJECT IDP DURABLE DCN: 2010-GEO-033 Date: August 30, 2012;Revised October 4, 2012 1 [Date] DISCLAIMER The author’s views expressed in this publication do not necessarily reflect the views of the Unit- ed States Agency for International Development or the United States Government 1 4 October 2012 Mr. Bradley Carr Water Irrigation and Infrastructure Advisor Office of Economic Growth US Agency for International Development 11 George Balanchine Street Tbilisi, 0131 Georgia Re: Environmental Scoping Statement for Oni Water Supply Dear Mr. Carr: This Scoping Statement is being submitted to you in accordance with the requirements of task order no. AID-114-TO-11-00002 of contract AID-EDH-I-00-08-00027-00. It provides Tetra Tech’s Environmental revised Scoping Statement for Oni Water Supply to address comments by BEO. We look forward to your review and welcome your comments and suggestions. Very truly yours, Jeffrey W. Fredericks, P.E., PhD Chief of Party Tetra Tech, Inc. USAID/ Caucasus – Municipal Infrastructure and IDP Housing Rehabilitation Project (GMIP) 10th Floor, 154 Aghmashenebeli Ave. Tbilisi, 0102, Georgia Tel: +995322910401, Fax: +995322910401 Email: [email protected] CC: USAID (George Kokochashvili); MDF (Kartlos Gviniashvili); Tetra Tech (Firouz Rooyani, Dean White, Brian Potvin) USAID/ Caucasus Municipal Infrastructure and IDP Housing Rehabilitation Project 10th Floor 154 Aghmashenebeli Ave. Tbilisi, -

Cryptic Speciation Patterns in Iranian Rock Lizards Uncovered by Integrative Taxonomy
Cryptic Speciation Patterns in Iranian Rock Lizards Uncovered by Integrative Taxonomy Faraham Ahmadzadeh1,2*, Morris Flecks2, Miguel A. Carretero3, Omid Mozaffari4, Wolfgang Bo¨ hme2,D. James Harris3, Susana Freitas3, Dennis Ro¨ dder2 1 Department of Biodiversity and Ecosystem Management, Environmental Sciences Research Institute, Shahid Beheshti University, Tehran, Iran, 2 Zoologisches Forschungsmuseum Alexander Koenig, Bonn, Germany, 3 Centro de Investigac¸a˜o em Biodiversidade e Recursos Gene´ticos, Universidade do Porto, Vaira˜o, Porto, Portugal, 4 Aria Herpetological Institute, Tehran, Iran Abstract While traditionally species recognition has been based solely on morphological differences either typological or quantitative, several newly developed methods can be used for a more objective and integrative approach on species delimitation. This may be especially relevant when dealing with cryptic species or species complexes, where high overall resemblance between species is coupled with comparatively high morphological variation within populations. Rock lizards, genus Darevskia, are such an example, as many of its members offer few diagnostic morphological features. Herein, we use a combination of genetic, morphological and ecological criteria to delimit cryptic species within two species complexes, D. chlorogaster and D. defilippii, both distributed in northern Iran. Our analyses are based on molecular information from two nuclear and two mitochondrial genes, morphological data (15 morphometric, 16 meristic and four categorical characters) and eleven newly calculated spatial environmental predictors. The phylogeny inferred for Darevskia confirmed monophyly of each species complex, with each of them comprising several highly divergent clades, especially when compared to other congeners. We identified seven candidate species within each complex, of which three and four species were supported by Bayesian species delimitation within D. -

Darevskia Praticola)
Amphibia-Reptilia 39 (2018): 229-238 Aspects of thermal ecology of the meadow lizard (Darevskia praticola) Jelena Corovi´ c´1,∗, Jelka Crnobrnja-Isailovic´1,2 Abstract. We studied the thermal biology of the meadow lizard (Darevskia praticola) in the peripheral part of its distribution range (westernmost edge of the distribution area). We assessed whether these lizards actively thermoregulate, estimated the accuracy and effectiveness of thermoregulation, and evaluated the thermal quality of the habitat using the standard thermal parameters: body (Tb), preferred (Tpref) with set-point range (Tset) and operative temperatures (Te). Tset of the meadow lizard under controlled laboratory conditions was between 27.8°C and 31.4°C. In the field Tb and Te averaged 29.0°C and 26.1°C, respectively. A large proportion of Tes fell below the Tset range of the meadow lizard, and lizard Tbs were substantially closer to the species’ Tset range. Obtained values of thermoregulatory indices suggested that the meadow lizard thermoregulated actively, with a rather high accuracy (db = 0.8) and effectiveness (E = 0.8andde − db = 2.6), and that their habitat at this locality was thermally favourable during the spring. Our results suggest that thermal requirements of the meadow lizard resemble those of alpine lacertids, while their TbsandTset are lower than in most lacertid lizards. Further thermoregulation studies could be an important step in predicting the impact of the global climate change on the meadow lizard and the risks of local extinctions of its peripheral populations. Keywords: field body temperatures, Lacertidae, peripheral populations, preferred temperatures, thermoregulation. Introduction According to Arnold (1987), European lacer- tid lizards do not differ much regarding feed- Reptiles thermoregulate in response to differ- ing ecology, foraging strategies, activity pat- ent temperatures, which enables them to gather terns and thermoregulatory behaviour. -

Status and Protection of Globally Threatened Species in the Caucasus
STATUS AND PROTECTION OF GLOBALLY THREATENED SPECIES IN THE CAUCASUS CEPF Biodiversity Investments in the Caucasus Hotspot 2004-2009 Edited by Nugzar Zazanashvili and David Mallon Tbilisi 2009 The contents of this book do not necessarily reflect the views or policies of CEPF, WWF, or their sponsoring organizations. Neither the CEPF, WWF nor any other entities thereof, assumes any legal liability or responsibility for the accuracy, completeness, or usefulness of any information, product or process disclosed in this book. Citation: Zazanashvili, N. and Mallon, D. (Editors) 2009. Status and Protection of Globally Threatened Species in the Caucasus. Tbilisi: CEPF, WWF. Contour Ltd., 232 pp. ISBN 978-9941-0-2203-6 Design and printing Contour Ltd. 8, Kargareteli st., 0164 Tbilisi, Georgia December 2009 The Critical Ecosystem Partnership Fund (CEPF) is a joint initiative of l’Agence Française de Développement, Conservation International, the Global Environment Facility, the Government of Japan, the MacArthur Foundation and the World Bank. This book shows the effort of the Caucasus NGOs, experts, scientific institutions and governmental agencies for conserving globally threatened species in the Caucasus: CEPF investments in the region made it possible for the first time to carry out simultaneous assessments of species’ populations at national and regional scales, setting up strategies and developing action plans for their survival, as well as implementation of some urgent conservation measures. Contents Foreword 7 Acknowledgments 8 Introduction CEPF Investment in the Caucasus Hotspot A. W. Tordoff, N. Zazanashvili, M. Bitsadze, K. Manvelyan, E. Askerov, V. Krever, S. Kalem, B. Avcioglu, S. Galstyan and R. Mnatsekanov 9 The Caucasus Hotspot N. -
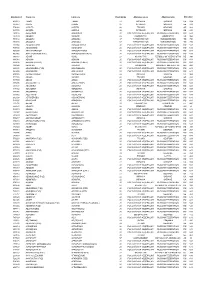
FÁK Állomáskódok
Állomáskód Orosz név Latin név Vasút kódja Államnév orosz Államnév latin Államkód 406513 1 МАЯ 1 MAIA 22 УКРАИНА UKRAINE UA 804 085827 ААКРЕ AAKRE 26 ЭСТОНИЯ ESTONIA EE 233 574066 ААПСТА AAPSTA 28 ГРУЗИЯ GEORGIA GE 268 085780 ААРДЛА AARDLA 26 ЭСТОНИЯ ESTONIA EE 233 269116 АБАБКОВО ABABKOVO 20 РОССИЙСКАЯ ФЕДЕРАЦИЯ RUSSIAN FEDERATION RU 643 737139 АБАДАН ABADAN 29 УЗБЕКИСТАН UZBEKISTAN UZ 860 753112 АБАДАН-I ABADAN-I 67 ТУРКМЕНИСТАН TURKMENISTAN TM 795 753108 АБАДАН-II ABADAN-II 67 ТУРКМЕНИСТАН TURKMENISTAN TM 795 535004 АБАДЗЕХСКАЯ ABADZEHSKAIA 20 РОССИЙСКАЯ ФЕДЕРАЦИЯ RUSSIAN FEDERATION RU 643 795736 АБАЕВСКИЙ ABAEVSKII 20 РОССИЙСКАЯ ФЕДЕРАЦИЯ RUSSIAN FEDERATION RU 643 864300 АБАГУР-ЛЕСНОЙ ABAGUR-LESNOI 20 РОССИЙСКАЯ ФЕДЕРАЦИЯ RUSSIAN FEDERATION RU 643 865065 АБАГУРОВСКИЙ (РЗД) ABAGUROVSKII (RZD) 20 РОССИЙСКАЯ ФЕДЕРАЦИЯ RUSSIAN FEDERATION RU 643 699767 АБАИЛ ABAIL 27 КАЗАХСТАН REPUBLIC OF KAZAKHSTAN KZ 398 888004 АБАКАН ABAKAN 20 РОССИЙСКАЯ ФЕДЕРАЦИЯ RUSSIAN FEDERATION RU 643 888108 АБАКАН (ПЕРЕВ.) ABAKAN (PEREV.) 20 РОССИЙСКАЯ ФЕДЕРАЦИЯ RUSSIAN FEDERATION RU 643 398904 АБАКЛИЯ ABAKLIIA 23 МОЛДАВИЯ MOLDOVA, REPUBLIC OF MD 498 889401 АБАКУМОВКА (РЗД) ABAKUMOVKA 20 РОССИЙСКАЯ ФЕДЕРАЦИЯ RUSSIAN FEDERATION RU 643 882309 АБАЛАКОВО ABALAKOVO 20 РОССИЙСКАЯ ФЕДЕРАЦИЯ RUSSIAN FEDERATION RU 643 408006 АБАМЕЛИКОВО ABAMELIKOVO 22 УКРАИНА UKRAINE UA 804 571706 АБАША ABASHA 28 ГРУЗИЯ GEORGIA GE 268 887500 АБАЗА ABAZA 20 РОССИЙСКАЯ ФЕДЕРАЦИЯ RUSSIAN FEDERATION RU 643 887406 АБАЗА (ЭКСП.) ABAZA (EKSP.) 20 РОССИЙСКАЯ ФЕДЕРАЦИЯ RUSSIAN FEDERATION RU 643 -

Herpetological Review
Herpetological Review Volume 41, Number 2 — June 2010 SSAR Offi cers (2010) HERPETOLOGICAL REVIEW President The Quarterly News-Journal of the Society for the Study of Amphibians and Reptiles BRIAN CROTHER Department of Biological Sciences Editor Southeastern Louisiana University ROBERT W. HANSEN Hammond, Louisiana 70402, USA 16333 Deer Path Lane e-mail: [email protected] Clovis, California 93619-9735, USA [email protected] President-elect JOSEPH MENDLELSON, III Zoo Atlanta, 800 Cherokee Avenue, SE Associate Editors Atlanta, Georgia 30315, USA e-mail: [email protected] ROBERT E. ESPINOZA KERRY GRIFFIS-KYLE DEANNA H. OLSON California State University, Northridge Texas Tech University USDA Forestry Science Lab Secretary MARION R. PREEST ROBERT N. REED MICHAEL S. GRACE PETER V. LINDEMAN USGS Fort Collins Science Center Florida Institute of Technology Edinboro University Joint Science Department The Claremont Colleges EMILY N. TAYLOR GUNTHER KÖHLER JESSE L. BRUNNER Claremont, California 91711, USA California Polytechnic State University Forschungsinstitut und State University of New York at e-mail: [email protected] Naturmuseum Senckenberg Syracuse MICHAEL F. BENARD Treasurer Case Western Reserve University KIRSTEN E. NICHOLSON Department of Biology, Brooks 217 Section Editors Central Michigan University Mt. Pleasant, Michigan 48859, USA Book Reviews Current Research Current Research e-mail: [email protected] AARON M. BAUER JOSHUA M. HALE BEN LOWE Department of Biology Department of Sciences Department of EEB Publications Secretary Villanova University MuseumVictoria, GPO Box 666 University of Minnesota BRECK BARTHOLOMEW Villanova, Pennsylvania 19085, USA Melbourne, Victoria 3001, Australia St Paul, Minnesota 55108, USA P.O. Box 58517 [email protected] [email protected] [email protected] Salt Lake City, Utah 84158, USA e-mail: [email protected] Geographic Distribution Geographic Distribution Geographic Distribution Immediate Past President ALAN M. -

Preliminary Analysis of Correlated Evolution of Morphology and Ecological Diversification in Lacertid Lizards
Butll. Soc. Cat. Herp., 19 (2011) Preliminary analysis of correlated evolution of morphology and ecological diversification in lacertid lizards Fèlix Amat Orriols Àrea d'Herpetologia, Museu de Granollers-Ciències Naturals. Francesc Macià 51. 08402 Granollers. Catalonia. Spain. [email protected] Resum S'ha investigat la diversitat morfològica en 129 espècies de lacèrtids i la seva relació amb l'ecologia, per mitjà de mètodes comparatius, utilitzant set variables morfomètriques. La mida corporal és la variable més important, determinant un gradient entre espècies de petita i gran mida independentment evolucionades al llarg de la filogènia dels lacèrtids. Aquesta variable està forta i positivament correlacionada amb les altres, emmascarant els patrons de diversitat morfològica. Anàlisis multivariants en les variables ajustades a la mida corporal mostren una covariació negativa entre les mides relatives de la cua i les extremitats. Remarcablement, les espècies arborícoles i semiarborícoles (Takydromus i el clade africà equatorial) han aparegut dues vegades independentment durant l'evolució dels lacèrtids i es caracteritzen per cues extremadament llargues i extremitats anteriors relativament llargues en comparació a les posteriors. El llangardaix arborícola i planador Holaspis, amb la seva cua curta, constitueix l’única excepció. Un altre cas de convergència ha estat trobat en algunes espècies que es mouen dins de vegetació densa o herba (Tropidosaura, Lacerta agilis, Takydromus amurensis o Zootoca) que presenten cues llargues i extremitats curtes. Al contrari, les especies que viuen en deserts, estepes o matollars amb escassa vegetació aïllada dins grans espais oberts han desenvolupat extremitats posteriors llargues i anteriors curtes per tal d'assolir elevades velocitats i maniobrabilitat. Aquest és el cas especialment de Acanthodactylus i Eremias Abstract Morphologic diversity was studied in 129 species of lacertid lizards and their relationship with ecology by means of comparative analysis on seven linear morphometric measurements. -

Ethnobiology of Georgia
SHOTA TUSTAVELI ZAAL KIKVIDZE NATIONAL SCIENCE FUNDATION ILIA STATE UNIVERSITY PRESS ETHNOBIOLOGY OF GEORGIA ISBN 978-9941-18-350-8 Tbilisi 2020 Ethnobiology of Georgia 2020 Zaal Kikvidze Preface My full-time dedication to ethnobiology started in 2012, since when it has never failed to fascinate me. Ethnobiology is a relatively young science with many blank areas still in its landscape, which is, perhaps, good motivation to write a synthetic text aimed at bridging the existing gaps. At this stage, however, an exhaustive representation of materials relevant to the ethnobiology of Georgia would be an insurmountable task for one author. My goal, rather, is to provide students and researchers with an introduction to my country’s ethnobiology. This book, therefore, is about the key traditions that have developed over a long history of interactions between humans and nature in Georgia, as documented by modern ethnobiologists. Acknowledgements: I am grateful to my colleagues – Rainer Bussmann, Narel Paniagua Zambrana, David Kikodze and Shalva Sikharulidze for the exciting and fruitful discussions about ethnobiology, and their encouragement for pushing forth this project. Rainer Bussmann read the early draft of this text and I am grateful for his valuable comments. Special thanks are due to Jana Ekhvaia, for her crucial contribution as project coordinator and I greatly appreciate the constant support from the staff and administration of Ilia State University. Finally, I am indebted to my fairy wordmother, Kate Hughes whose help was indispensable at the later stages of preparation of this manuscript. 2 Table of contents Preface.......................................................................................................................................................... 2 Chapter 1. A brief introduction to ethnobiology...................................................................................... -

A Description of a New Subspecies of Rock Lizard Darevskia Brauneri Myusserica Ssp
Труды Зоологического института РАН Том 315, № 3, 2011, c. 242–262 УДК 598.113.6 ОПИСАНИЕ НОВОГО ПОДВИДА СКАЛЬНОЙ ЯЩЕРИЦЫ DAREVSKIA BRAUNERI MYUSSERICA SSP. NOV. ИЗ ЗАПАДНОГО ЗАКАВКАЗЬЯ (АБХАЗИЯ) C КОММЕНТАРИЯМИ ПО СИСТЕ МАТИКЕ КОМПЛЕКСА DAREVSKIA SAXICOLA И.В. Доронин Зоологический институт Российской академии наук, Университетская наб. 1, 199034 Санкт-Петербург, Россия; e-mail: [email protected] РЕЗЮМЕ В статье приводится описание нового подвида скальной ящерицы комплекса Darevskia saxicola, обитающего на территории Пицундо-Мюссерского заповедника и в районе г. Гагра Республики Абхазия. Мюссерская ящерица, Darevskia brauneri myusserica ssp. nov., отличается от других таксонов комплекса следующей ком- бинацией морфологических признаков: (1) крупный или очень крупный центральновисочный щиток; (2) прерывистый ряд ресничных зернышек между верхнересничными и надглазничными щитками; (3) наличие дополнительных щитков, лежащих по обе стороны от затылочного и межтеменного щитков, либо дробление последнего; (4) сетчатый рисунок на спине (у самок нечеткий); (5) доминирование у самок серого и светло- серого цвета в окраске дорсальной поверхности тела; (6) белое горло и брюхо. Кроме того, новый подвид отличается некоторыми особенностями биологии: биотопической приуроченностью к прибрежным выходам конгломерата и относительно низкой численностью популяции. Предположительно, формирование таксона протекало в плейстоцене. Образование приморской равнины полуострова Пицунда за счет аллювиальной и морской аккумуляции в позднем неоплейстоцене – голоцене разделило ареал мюссерской ящерицы на гагр- ский и мюссерский участки. Эта территория расположена в пределах Черноморского рефугиума восточно- средиземноморских видов герпетофауны. Ключевые слова: Абхазия, комплекс Darevskia saxicola, скальные ящерицы, Darevskia brauneri myusserica ssp. nov. A DESCRIPTION OF A NEW SUBSPECIES OF ROCK LIZARD DAREVSKIA BRAUNERI MYUSSERICA SSP. NOV. FROM THE WESTERN TRANSCAUCASIA (ABKHAZIA), WITH COMMENTS ON SYSTEMATICS OF DAREVSKIA SAXICOLA COMPLEX I.V. -

Literature Cited in Lizards Natural History Database
Literature Cited in Lizards Natural History database Abdala, C. S., A. S. Quinteros, and R. E. Espinoza. 2008. Two new species of Liolaemus (Iguania: Liolaemidae) from the puna of northwestern Argentina. Herpetologica 64:458-471. Abdala, C. S., D. Baldo, R. A. Juárez, and R. E. Espinoza. 2016. The first parthenogenetic pleurodont Iguanian: a new all-female Liolaemus (Squamata: Liolaemidae) from western Argentina. Copeia 104:487-497. Abdala, C. S., J. C. Acosta, M. R. Cabrera, H. J. Villaviciencio, and J. Marinero. 2009. A new Andean Liolaemus of the L. montanus series (Squamata: Iguania: Liolaemidae) from western Argentina. South American Journal of Herpetology 4:91-102. Abdala, C. S., J. L. Acosta, J. C. Acosta, B. B. Alvarez, F. Arias, L. J. Avila, . S. M. Zalba. 2012. Categorización del estado de conservación de las lagartijas y anfisbenas de la República Argentina. Cuadernos de Herpetologia 26 (Suppl. 1):215-248. Abell, A. J. 1999. Male-female spacing patterns in the lizard, Sceloporus virgatus. Amphibia-Reptilia 20:185-194. Abts, M. L. 1987. Environment and variation in life history traits of the Chuckwalla, Sauromalus obesus. Ecological Monographs 57:215-232. Achaval, F., and A. Olmos. 2003. Anfibios y reptiles del Uruguay. Montevideo, Uruguay: Facultad de Ciencias. Achaval, F., and A. Olmos. 2007. Anfibio y reptiles del Uruguay, 3rd edn. Montevideo, Uruguay: Serie Fauna 1. Ackermann, T. 2006. Schreibers Glatkopfleguan Leiocephalus schreibersii. Munich, Germany: Natur und Tier. Ackley, J. W., P. J. Muelleman, R. E. Carter, R. W. Henderson, and R. Powell. 2009. A rapid assessment of herpetofaunal diversity in variously altered habitats on Dominica. -

Дифференциация И Систематика Скальных Ящериц Комплекса Darevskia (Saxicola) (Sauria: Lacertidae) По Данным Морфологического И Молекулярного Анализов
Труды Зоологического института РАН Том 317, № 1, 2013, c. 54–84 УДК 575+598.1 ДИФФЕРЕНЦИАЦИЯ И СИСТЕМАТИКА СКАЛЬНЫХ ЯЩЕРИЦ КОМПЛЕКСА DAREVSKIA (SAXICOLA) (SAURIA: LACERTIDAE) ПО ДАННЫМ МОРФОЛОГИЧЕСКОГО И МОЛЕКУЛЯРНОГО АНАЛИЗОВ И.В. Доронин1*, Б.С. Туниев2 и О.В. Кукушкин3 1Зоологический институт Российской академии наук, Россия, 199034, Санкт-Петербург, Университетская наб. 1, e-mail: [email protected] 2Сочинский национальный парк, Россия, 354000, Краснодарский край, Сочи, ул. Московская, 21, e-mail: [email protected] 3Карадагский природный заповедник НАН Украины, Украина, 98188, автономная республика Крым, Феодосия, пгт. Курортное, ул. Науки, 24, e-mail: [email protected] РЕЗЮМЕ Результаты статистического анализа морфологических признаков шести распространенных на Кавказе и в Крыму форм скальных ящериц комплекса Darevskia (saxicola), наряду с данными исследования изменчиво- сти фрагмента гена цитохром b митохондриальной ДНК, указывают на глубокую дифференциацию внутри этого комплекса, говорят в пользу видовой самостоятельности D. szczerbaki (Lukina, 1963) и обособленно- сти недавно описанного таксона D. brauneri myusserica Doronin, 2011. Вместе с тем внутривидовая измен- чивость D. brauneri (Méhely, 1909) позволяет выделить только две валидные формы подвидового статуса: D. b. brauneri и D. b. myusserica. По нашему мнению, Lacerta saxicola darevskii Szczerbak, 1962 (= D. brauneri darevskii) должна рассматриваться как младший синоним D. b. brauneri. Обсуждаются возможные сценарии происхождения форм комплекса. Ключевые слова: комплекс -

6. Imereti – Historical-Cultural Overview
SFG2110 SECOND REGIONAL DEVELOPMETN PROJECT IMERETI REGIONAL DEVELOPMENT PROGRAM IMERETI TOURISM DEVELOPMENT STRATEGY Public Disclosure Authorized STRATEGIC ENVIRONMENTAL, CULTURAL HERITAGE AND SOCIAL ASSESSMENT Public Disclosure Authorized Public Disclosure Authorized Public Disclosure Authorized Tbilisi, December, 2014 ABBREVIATIONS GNTA Georgia National Tourism Administration EIA Environnemental Impact Assessment EMP Environmental Management Plan EMS Environmental Management System IFI International Financial Institution IRDS Imereti Regional Development Strategy ITDS Imereti Tourism Development Strategy MDF Municipal Development Fund of Georgia MoA Ministry of Agriculture MoENRP Ministry of Environment and Natural Resources Protection of Georgia MoIA Ministry of Internal Affairs MoCMP Ministry of Culture and Monument Protection MoJ Ministry of Justice MoESD Ministry of Economic and Sustaineble Developmnet NACHP National Agency for Cultural Heritage Protection PIU Project Implementation Unit PPE Personal protective equipment RDP Regional Development Project SECHSA Strategic Environmental, Cultural Heritage and Social Assessment WB World Bank Contents EXECUTIVE SUMMARY ........................................................................................................................................... 0 1. INTRODUCTION ........................................................................................................................................... 14 1.1 PROJECT CONTEXT ...............................................................................................................................