Systemic Vitamin Intake Impacting Tissue Proteomes Heesoo Jeong and Nathaniel M
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Boards' Fodder
boards’ fodder Cosmeceuticals Contributed by Elisabeth Hurliman, MD, PhD; Jennifer Hayes, MD; Hilary Reich MD; and Sarah Schram, MD. INGREDIENT FUNCTION MECHANISM ASSOCIATIONS/SIDE EFFECTS Vitamin A/ Antioxidant (reduces free Affects gene transcription Comedolysis epidermal thickening, dermal Derivatives (retinal, radicals, lowers concentration differentiation and growth of regeneration, pigment lightening retinol, retinoic of matrix metalloproteinases cells in the skin acid, provitamin reduces collagen degradation) Side effects: Irritation, erythema, desquamation A, asthaxanthin, Normalizes follicular Elisabeth Hurliman, lutein) epithelial differentiation and keratinization MD, PhD, is a PGY-4 dermatology resident Vitamin C (L Secondary endogenous Ascorbic acid: necessary L-ascorbic acid + alpha-tocopherol (vitamin E)= ascorbic acid, antioxidant in skin cofactor for prolylhydroxylase UVA and UVB protection at University of tetrahexyldecyl and lysyl hydroxylase Minnesota department ascorbate) Lightens pigment Zinc, resveratrol, L-ergothioneine and tyrosine add of dermatology. (affects melanogenesis) L-ascorbic acid: scavenges to vitamin C bioavailability free oxygen radicals, Protects Vitamin E from oxidation stimulates collagen synthesis Improves skin texture and hydration May interrupt melanogenesis by interacting with copper ions Vitamin E/ Primary endogenous antioxidant Prevents lipid peroxidation; Alpha tocopherol is the most physiologically Tocopherols, in skin scavenges free oxygen active isomer Jennifer Hayes, MD, Tocotrienols -

Lactobacillus Acidophilus Nucelic Acid Sequences Encoding Carbohydrate Utilization-Related Proteins and Uses Therefor
(19) & (11) EP 2 407 481 A1 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 18.01.2012 Bulletin 2012/03 C07K 14/335 (2006.01) C12N 15/31 (2006.01) C12N 15/54 (2006.01) C12N 9/12 (2006.01) (2006.01) (2006.01) (21) Application number: 11176351.2 C12N 15/74 C12Q 1/37 (22) Date of filing: 08.03.2005 (84) Designated Contracting States: • Russel, Michael W. AT BE BG CH CY CZ DE DK EE ES FI FR GB GR Newburgh, IN 47630 (US) HU IE IS IT LI LT LU MC NL PL PT RO SE SI SK TR • Duong, Tri Raleigh, North Carolina 27606 (US) (30) Priority: 08.03.2004 US 551121 P 07.03.2005 US 74226 (74) Representative: Williams, Aylsa D Young & Co LLP (62) Document number(s) of the earlier application(s) in 120 Holborn accordance with Art. 76 EPC: London EC1N 2DY (GB) 10196268.6 / 2 348 041 05760391.2 / 1 727 826 Remarks: •Thecomplete document including Reference Tables (71) Applicant: North Carolina State University and the Sequence Listing can be downloaded from Raleigh, North Carolina 27606 (US) the EPO website •This application was filed on 02-08-2011 as a (72) Inventors: divisional application to the application mentioned • Klaenhammer, Todd Robert under INID code 62. Raleigh, NC 27606 (US) •Claims filed after the date of filing of the application/ • Altermann, Eric after the date of receipt of the divisional application Apex, NC 27539 (US) (Rule 68(4) EPC). • Barrangou, Rodolphe Madison, WI 53718 (US) (54) Lactobacillus acidophilus nucelic acid sequences encoding carbohydrate utilization-related proteins and uses therefor (57) Carbohydrate utilization-related and multidrug invention also provides vectors containing a nucleic acid transporter nucleic acids and polypeptides, and frag- of the invention and cells into which the vector has been ments and variants therof, are disclosed in the current introduced. -

Overview of Bariatric Surgery for the Physician
■ CLINICAL PRACTICE Clinical Medicine 2012, Vol 12, No 5: 435–40 Overview of bariatric surgery for the physician Keng Ngee Hng and Yeng S Ang ABSTRACT – The worldwide pandemic of obesity carries effectiveness2,6,15 have fuelled an increase in the number of pro- alarming health and socioeconomic implications. Bariatric cedures performed. surgery is currently the only effective treatment for severe obesity. It is safe, with mortality comparable to that of chole- Types of surgery cystectomy, and effective in producing substantial and sus- tainable weight loss, along with high rates of resolution of Bariatric surgical procedures are traditionally classified as restric- associated comorbidities, including type 2 diabetes. For this tive, malabsorptive or combined according to their mechanism reason, indications for bariatric surgery are being widened. In of action. The procedures most commonly performed are addition to volume restriction and malabsorption, bariatric laparoscopic adjustable gastric banding and roux-en-y gastric surgery brings about neurohormonal changes that affect bypass.3,13 Sleeve gastrectomy is increasingly performed.2,6,7 satiety and glucose homeostasis. Increased understanding of Biliopancreatic diversion and biliopancreatic diversion with these mechanisms will help realise therapeutic benefits by duodenal switch are much more complex and performed infre- pharmacological means. Bariatric surgery improves long-term quently.2,5,17,22 Other historical procedures are no longer in mortality but can cause long-term nutritional deficiencies. common use. The safety of pregnancy after bariatric surgery is still being In addition to restriction and malabsorption, recent evidence elucidated. suggests that neurohormonal changes are an important effect of bariatric surgery.2,6,7,17,18 Bariatric surgery is only part of the KEY WORDS: bariatric surgery, obesity, weight loss, diabetes, management of severe obesity. -

Metabolism of Levulinate and Conversion to the Drug Of
METABOLISM OF LEVULINATE AND CONVERSION TO THE DRUG OF ABUSE 4-HYDROXYPENTANOATE by STEPHANIE R. HARRIS Submitted in partial fulfillment of the requirements For the Degree of Doctor of Philosophy Thesis Advisor: Henri Brunengraber, M.D., Ph.D. Department of Nutrition CASE WESTERN RESERVE UNIVERSITY August 2011 CASE WESTERN RESERVE UNIVERSITY SCHOOL OF GRADUATE STUDIES We hereby approve the thesis/dissertation of STEPHANIE R. HARRIS candidate for the ________________________________ Doctor of Philosophy degree *. (signed) ________________________________________________ Edith Lerner, PhD (Chair of the committee) ________________________________________________ Henri Brunengraber, MD, PhD ________________________________________________ Colleen Croniger, PhD ________________________________________________ Paul Ernsberger, PhD ________________________________________________ Janos Kerner, PhD ________________________________________________ Michelle Puchowicz, PhD (date) _______________________June 16, 2011 *We also certify that written approval has been obtained for any proprietary material contained therein. ii DEDICATION I dedicate this work to my parents and to my husband, Paul. My parents have provided continuous love, encouragement, and guidance throughout my life. They have taught me to set my goals high. My husband has been a source of strength and inspiration, and his dedication and enthusiastic support have helped me achieve this work. iii TABLE OF CONTENTS Table of Contents………………………………………………………………………... iv List of Tables…………………………………………………………………………....viii -

Molybdoproteomes and Evolution of Molybdenum Utilization
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Vadim Gladyshev Publications Biochemistry, Department of April 2008 Molybdoproteomes and evolution of molybdenum utilization Yan Zhang University of Nebraska-Lincoln, [email protected] Vadim N. Gladyshev University of Nebraska-Lincoln, [email protected] Follow this and additional works at: https://digitalcommons.unl.edu/biochemgladyshev Part of the Biochemistry, Biophysics, and Structural Biology Commons Zhang, Yan and Gladyshev, Vadim N., "Molybdoproteomes and evolution of molybdenum utilization" (2008). Vadim Gladyshev Publications. 78. https://digitalcommons.unl.edu/biochemgladyshev/78 This Article is brought to you for free and open access by the Biochemistry, Department of at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Vadim Gladyshev Publications by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Published in Journal of Molecular Biology (2008); doi: 10.1016/j.jmb.2008.03.051 Copyright © 2008 Elsevier. Used by permission. http://www.sciencedirect.com/science/journal/00222836 Submitted November 26, 2007; revised March 15, 2008; accepted March 25, 2008; published online as “Accepted Manuscript” April 1, 2008. Molybdoproteomes and evolution of molybdenum utilization Yan Zhang and Vadim N. Gladyshev* Department of Biochemistry, University of Nebraska–Lincoln, Lincoln, NE 685880664 *Corresponding author—tel 402 472-4948, fax 402 472-7842, email [email protected] Abstract The trace element molybdenum (Mo) is utilized in many life forms, where it is a key component of several enzymes involved in nitrogen, sulfur, and carbon metabolism. With the exception of nitrogenase, Mo is bound in proteins to a pterin, thus forming the molybdenum cofactor (Moco) at the catalytic sites of molybdoenzymes. -

Supplement 1 Overview of Dystonia Genes
Supplement 1 Overview of genes that may cause dystonia in children and adolescents Gene (OMIM) Disease name/phenotype Mode of inheritance 1: (Formerly called) Primary dystonias (DYTs): TOR1A (605204) DYT1: Early-onset generalized AD primary torsion dystonia (PTD) TUBB4A (602662) DYT4: Whispering dystonia AD GCH1 (600225) DYT5: GTP-cyclohydrolase 1 AD deficiency THAP1 (609520) DYT6: Adolescent onset torsion AD dystonia, mixed type PNKD/MR1 (609023) DYT8: Paroxysmal non- AD kinesigenic dyskinesia SLC2A1 (138140) DYT9/18: Paroxysmal choreoathetosis with episodic AD ataxia and spasticity/GLUT1 deficiency syndrome-1 PRRT2 (614386) DYT10: Paroxysmal kinesigenic AD dyskinesia SGCE (604149) DYT11: Myoclonus-dystonia AD ATP1A3 (182350) DYT12: Rapid-onset dystonia AD parkinsonism PRKRA (603424) DYT16: Young-onset dystonia AR parkinsonism ANO3 (610110) DYT24: Primary focal dystonia AD GNAL (139312) DYT25: Primary torsion dystonia AD 2: Inborn errors of metabolism: GCDH (608801) Glutaric aciduria type 1 AR PCCA (232000) Propionic aciduria AR PCCB (232050) Propionic aciduria AR MUT (609058) Methylmalonic aciduria AR MMAA (607481) Cobalamin A deficiency AR MMAB (607568) Cobalamin B deficiency AR MMACHC (609831) Cobalamin C deficiency AR C2orf25 (611935) Cobalamin D deficiency AR MTRR (602568) Cobalamin E deficiency AR LMBRD1 (612625) Cobalamin F deficiency AR MTR (156570) Cobalamin G deficiency AR CBS (613381) Homocysteinuria AR PCBD (126090) Hyperphelaninemia variant D AR TH (191290) Tyrosine hydroxylase deficiency AR SPR (182125) Sepiaterine reductase -

Enzymatic Encoding Methods for Efficient Synthesis Of
(19) TZZ__T (11) EP 1 957 644 B1 (12) EUROPEAN PATENT SPECIFICATION (45) Date of publication and mention (51) Int Cl.: of the grant of the patent: C12N 15/10 (2006.01) C12Q 1/68 (2006.01) 01.12.2010 Bulletin 2010/48 C40B 40/06 (2006.01) C40B 50/06 (2006.01) (21) Application number: 06818144.5 (86) International application number: PCT/DK2006/000685 (22) Date of filing: 01.12.2006 (87) International publication number: WO 2007/062664 (07.06.2007 Gazette 2007/23) (54) ENZYMATIC ENCODING METHODS FOR EFFICIENT SYNTHESIS OF LARGE LIBRARIES ENZYMVERMITTELNDE KODIERUNGSMETHODEN FÜR EINE EFFIZIENTE SYNTHESE VON GROSSEN BIBLIOTHEKEN PROCEDES DE CODAGE ENZYMATIQUE DESTINES A LA SYNTHESE EFFICACE DE BIBLIOTHEQUES IMPORTANTES (84) Designated Contracting States: • GOLDBECH, Anne AT BE BG CH CY CZ DE DK EE ES FI FR GB GR DK-2200 Copenhagen N (DK) HU IE IS IT LI LT LU LV MC NL PL PT RO SE SI • DE LEON, Daen SK TR DK-2300 Copenhagen S (DK) Designated Extension States: • KALDOR, Ditte Kievsmose AL BA HR MK RS DK-2880 Bagsvaerd (DK) • SLØK, Frank Abilgaard (30) Priority: 01.12.2005 DK 200501704 DK-3450 Allerød (DK) 02.12.2005 US 741490 P • HUSEMOEN, Birgitte Nystrup DK-2500 Valby (DK) (43) Date of publication of application: • DOLBERG, Johannes 20.08.2008 Bulletin 2008/34 DK-1674 Copenhagen V (DK) • JENSEN, Kim Birkebæk (73) Proprietor: Nuevolution A/S DK-2610 Rødovre (DK) 2100 Copenhagen 0 (DK) • PETERSEN, Lene DK-2100 Copenhagen Ø (DK) (72) Inventors: • NØRREGAARD-MADSEN, Mads • FRANCH, Thomas DK-3460 Birkerød (DK) DK-3070 Snekkersten (DK) • GODSKESEN, -

Role of Neuronal Glucosensing in the Regulation of Energy Homeostasis Barry E
Role of Neuronal Glucosensing in the Regulation of Energy Homeostasis Barry E. Levin,1,2 Ling Kang,2 Nicole M. Sanders,3 and Ambrose A. Dunn-Meynell1,2 Glucosensing is a property of specialized neurons in the studies of damage to the hypothalamus pointed to the brain that regulate their membrane potential and firing brain as the primary regulator of energy homeostasis. rate as a function of ambient glucose levels. These neurons Lesions of the ventromedial hypothalamus (VMH) produce have several similarities to - and ␣-cells in the pancreas, increased food intake (hyperphagia), obesity (1), and which are also responsive to ambient glucose levels. Many defective autonomic function in organs involved in the use glucokinase as a rate-limiting step in the production of ATP and its effects on membrane potential and ion channel regulation of energy expenditure (2,3). On the other hand, function to sense glucose. Glucosensing neurons are orga- electrical stimulation of the VMH leads to generalized nized in an interconnected distributed network throughout sympathoadrenal activation (4) with increased activity in the brain that also receives afferent neural input from thermogenic tissues (5). Lesions of the lateral hypotha- glucosensors in the liver, carotid body, and small intes- lamic area (LHA) reduce food intake and increase sympa- tines. In addition to glucose, glucosensing neurons can use thetic activity and eventually establish a new lower other metabolic substrates, hormones, and peptides to defended body weight (3,5,6). Whereas such early studies regulate their firing rate. Consequently, the output of pointed to the hypothalamus as the central controller of these “metabolic sensing” neurons represents their in- tegrated response to all of these simultaneous inputs. -

Abstracts from the 9Th Biennial Scientific Meeting of The
International Journal of Pediatric Endocrinology 2017, 2017(Suppl 1):15 DOI 10.1186/s13633-017-0054-x MEETING ABSTRACTS Open Access Abstracts from the 9th Biennial Scientific Meeting of the Asia Pacific Paediatric Endocrine Society (APPES) and the 50th Annual Meeting of the Japanese Society for Pediatric Endocrinology (JSPE) Tokyo, Japan. 17-20 November 2016 Published: 28 Dec 2017 PS1 Heritable forms of primary bone fragility in children typically lead to Fat fate and disease - from science to global policy a clinical diagnosis of either osteogenesis imperfecta (OI) or juvenile Peter Gluckman osteoporosis (JO). OI is usually caused by dominant mutations affect- Office of Chief Science Advsor to the Prime Minister ing one of the two genes that code for two collagen type I, but a re- International Journal of Pediatric Endocrinology 2017, 2017(Suppl 1):PS1 cessive form of OI is present in 5-10% of individuals with a clinical diagnosis of OI. Most of the involved genes code for proteins that Attempts to deal with the obesity epidemic based solely on adult be- play a role in the processing of collagen type I protein (BMP1, havioural change have been rather disappointing. Indeed the evidence CREB3L1, CRTAP, LEPRE1, P4HB, PPIB, FKBP10, PLOD2, SERPINF1, that biological, developmental and contextual factors are operating SERPINH1, SEC24D, SPARC, from the earliest stages in development and indeed across generations TMEM38B), or interfere with osteoblast function (SP7, WNT1). Specific is compelling. The marked individual differences in the sensitivity to the phenotypes are caused by mutations in SERPINF1 (recessive OI type obesogenic environment need to be understood at both the individual VI), P4HB (Cole-Carpenter syndrome) and SEC24D (‘Cole-Carpenter and population level. -

Contig Protein Description Symbol Anterior Posterior Ratio
Table S2. List of proteins detected in anterior and posterior intestine pooled samples. Data on protein expression are mean ± SEM of 4 pools fed the experimental diets. The number of the contig in the Sea Bream Database (http://nutrigroup-iats.org/seabreamdb) is indicated. Contig Protein Description Symbol Anterior Posterior Ratio Ant/Pos C2_6629 1,4-alpha-glucan-branching enzyme GBE1 0.88±0.1 0.91±0.03 0.98 C2_4764 116 kDa U5 small nuclear ribonucleoprotein component EFTUD2 0.74±0.09 0.71±0.05 1.03 C2_299 14-3-3 protein beta/alpha-1 YWHAB 1.45±0.23 2.18±0.09 0.67 C2_268 14-3-3 protein epsilon YWHAE 1.28±0.2 2.01±0.13 0.63 C2_2474 14-3-3 protein gamma-1 YWHAG 1.8±0.41 2.72±0.09 0.66 C2_1017 14-3-3 protein zeta YWHAZ 1.33±0.14 4.41±0.38 0.30 C2_34474 14-3-3-like protein 2 YWHAQ 1.3±0.11 1.85±0.13 0.70 C2_4902 17-beta-hydroxysteroid dehydrogenase 14 HSD17B14 0.93±0.05 2.33±0.09 0.40 C2_3100 1-acylglycerol-3-phosphate O-acyltransferase ABHD5 ABHD5 0.85±0.07 0.78±0.13 1.10 C2_15440 1-phosphatidylinositol phosphodiesterase PLCD1 0.65±0.12 0.4±0.06 1.65 C2_12986 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase delta-1 PLCD1 0.76±0.08 1.15±0.16 0.66 C2_4412 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma-2 PLCG2 1.13±0.08 2.08±0.27 0.54 C2_3170 2,4-dienoyl-CoA reductase, mitochondrial DECR1 1.16±0.1 0.83±0.03 1.39 C2_1520 26S protease regulatory subunit 10B PSMC6 1.37±0.21 1.43±0.04 0.96 C2_4264 26S protease regulatory subunit 4 PSMC1 1.2±0.2 1.78±0.08 0.68 C2_1666 26S protease regulatory subunit 6A PSMC3 1.44±0.24 1.61±0.08 -

Nutrition 102 – Class 3
Nutrition 102 – Class 3 Angel Woolever, RD, CD 1 Nutrition 102 “Introduction to Human Nutrition” second edition Edited by Michael J. Gibney, Susan A. Lanham-New, Aedin Cassidy, and Hester H. Vorster May be purchased online but is not required for the class. 2 Technical Difficulties Contact: Erin Deichman 574.753.1706 [email protected] 3 Questions You may raise your hand and type your question. All questions will be answered at the end of the webinar to save time. 4 Review from Last Week Vitamins E, K, and C What it is Source Function Requirement Absorption Deficiency Toxicity Non-essential compounds Bioflavonoids: Carnitine, Choline, Inositol, Taurine, and Ubiquinone Phytoceuticals 5 Priorities for Today’s Session B Vitamins What they are Source Function Requirement Absorption Deficiency Toxicity 6 7 What Is Vitamin B1 First B Vitamin to be discovered 8 Vitamin B1 Sources Pork – rich source Potatoes Whole-grain cereals Meat Fish 9 Functions of Vitamin B1 Converts carbohydrates into glucose for energy metabolism Strengthens immune system Improves body’s ability to withstand stressful conditions 10 Thiamine Requirements Groups: RDA (mg/day): Infants 0.4 Children 0.7-1.2 Males 1.5 Females 1 Pregnancy 2 Lactation 2 11 Thiamine Absorption Absorbed in the duodenum and proximal jejunum Alcoholics are especially susceptible to thiamine deficiency Excreted in urine, diuresis, and sweat Little storage of thiamine in the body 12 Barriers to Thiamine Absorption Lost into cooking water Unstable to light Exposure to sunlight Destroyed -
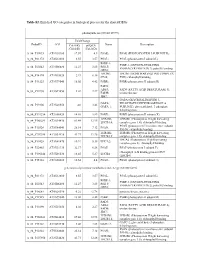
Table S2. Enriched GO Categories in Biological Process for the Shared Degs
Table S2. Enriched GO categories in biological process for the shared DEGs photosynthesis (GO ID:15979) Fold Change ProbeID AGI Col-0(R) pifQ(D) Name Description /Col-0(D) /Col-0(D) A_84_P19035 AT1G30380 17.07 4.9 PSAK; PSAK (PHOTOSYSTEM I SUBUNIT K) A_84_P21372 AT4G12800 8.55 3.57 PSAL; PSAL (photosystem I subunit L) PSBP-1; PSBP-1 (OXYGEN-EVOLVING A_84_P20343 AT1G06680 12.27 3.85 PSII-P; ENHANCER PROTEIN 2); poly(U) binding OEE2; LHCB6; LHCB6 (LIGHT HARVESTING COMPLEX A_84_P14174 AT1G15820 23.9 6.16 CP24; PSII); chlorophyll binding A_84_P11525 AT1G79040 16.02 4.42 PSBR; PSBR (photosystem II subunit R) FAD5; ADS3; FAD5 (FATTY ACID DESATURASE 5); A_84_P19290 AT3G15850 4.02 2.27 FADB; oxidoreductase JB67; GAPA (GLYCERALDEHYDE 3- GAPA; PHOSPHATE DEHYDROGENASE A A_84_P19306 AT3G26650 4.6 3.43 GAPA-1; SUBUNIT); glyceraldehyde-3-phosphate dehydrogenase A_84_P193234 AT2G06520 14.01 3.89 PSBX; PSBX (photosystem II subunit X) LHB1B1; LHB1B1 (Photosystem II light harvesting A_84_P160283 AT2G34430 89.44 32.95 LHCB1.4; complex gene 1.4); chlorophyll binding PSAN (photosystem I reaction center subunit A_84_P10324 AT5G64040 26.14 7.12 PSAN; PSI-N); calmodulin binding LHB1B2; LHB1B2 (Photosystem II light harvesting A_84_P207958 AT2G34420 41.71 12.26 LHCB1.5; complex gene 1.5); chlorophyll binding LHCA2 (Photosystem I light harvesting A_84_P19428 AT3G61470 10.91 5.36 LHCA2; complex gene 2); chlorophyll binding A_84_P22465 AT1G31330 32.37 6.58 PSAF; PSAF (photosystem I subunit F) chlorophyll A-B binding protein CP29 A_84_P190244 AT5G01530 16.45 5.27 LHCB4