Site-Directed Amino Acid Substitutions in the Hydroxylase
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Formation of Butane in the Polymerization of Methyl Methacrylate with Butyllithium
Polymer Journal, Vol. 12, No. 8, pp 535-537 (1980) SHORT COMMUNICATION The Formation of Butane in the Polymerization of Methyl Methacrylate with Butyllithium Koichi HATADA, Tatsuki KITAYAMA, and Heimei YUKI Department of Chemistry, Faculty of Engineering Science, Osaka University, Machikaneyama, Toyonaka, Osaka 560, Japan. (Received February 28, 1980) KEY WORDS Methyl Methacrylate I Perdeuterated Methyl Methacrylate I Anionic Polymerization I Butyllithium I Toluene I Tetrahydrofuran I Butane Formation I Mass Spectroscopy I Butene I It has been found that a small amount of butane merization reactions in toluene: (1) the poly is formed when the polymerization of methyl merization of MMA terminated by CH30D methacrylate is initiated with butyllithium and (MMA-CH30D system) and (2) that of MMA-d8 terminated with acetic acid. The origin of the butane terminated by CH3 0H (MMA-d8-CH3 0H system). was not clear, although the proton abstraction from The reactions were quenched at 10 min following the monomer is assumed. 1 the initiation. The spectra of butane (BuH) and In this work the polymerizations of undeuterated butane-d-1, CH3 CH2 CH2CH2 D, (BuD) are also (MMA) and perdeuterated (MMA-d8 ) methyl shown for comparison. The BuH and BuD were methacrylates were initiated with undeuterated obtained by the reaction of BuLi with CH30H and butyllithium (BuLi) and terminated by CH3 0D and CH30D, respectively. By inspecting these four CH3 0H, respectively. Deuterium distributions in spectra, it can be seen that the butane from the the butane formed were determined by combined MMA-CH3 0D system mainly consists of BuD and gas-liquid phase chromatography-mass spectrom the butane from the MMA-d8-CH30H system is etry. -

Safety Data Sheet
SAFETY DATA SHEET 1. Identification Product number 1000007900 Product identifier AUTO GLASS QUICK RELEASE AGENT Company information Sprayway, Inc. 1005 S. Westgate Drive Addison, IL 60101 United States Company phone General Assistance 1-630-628-3000 Emergency telephone US 1-866-836-8855 Emergency telephone outside 1-952-852-4646 US Version # 01 Recommended use Not available. Recommended restrictions None known. 2. Hazard(s) identification Physical hazards Flammable aerosols Category 1 Health hazards Serious eye damage/eye irritation Category 2A Specific target organ toxicity, single exposure Category 3 narcotic effects Aspiration hazard Category 1 Environmental hazards Not classified. OSHA defined hazards Not classified. Label elements Signal word Danger Hazard statement Extremely flammable aerosol. May be fatal if swallowed and enters airways. Causes serious eye irritation. May cause drowsiness or dizziness. Precautionary statement Prevention Keep away from heat/sparks/open flames/hot surfaces. - No smoking. Do not spray on an open flame or other ignition source. Pressurized container: Do not pierce or burn, even after use. Avoid breathing gas. Wash thoroughly after handling. Use only outdoors or in a well-ventilated area. Wear eye/face protection. Response If swallowed: Immediately call a poison center/doctor. If inhaled: Remove person to fresh air and keep comfortable for breathing. If in eyes: Rinse cautiously with water for several minutes. Remove contact lenses, if present and easy to do. Continue rinsing. Call a poison center/doctor if you feel unwell. Do NOT induce vomiting. If eye irritation persists: Get medical advice/attention. Storage Store in a well-ventilated place. Keep container tightly closed. Store locked up. -
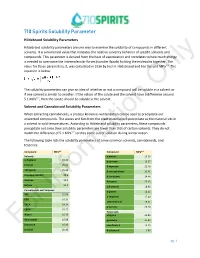
710 Spirits Solubility Parameter Hildebrand Solubility Parameters
710 Spirits Solubility Parameter Hildebrand Solubility Parameters Hildebrand solubility parameters are one way to examine the solubility of compounds in different solvents. It is a numerical value that indicates the relative solvency behavior of specific solvents and compounds. This parameter is derived from the heat of vaporization and correlates to how much energy is needed to overcome the intermolecular forces (van der Waals) holding the molecules together. The value for these parameters, δ, was calculated in 1936 by Joel H. Hildebrand and has the unit MPa1/2. The equation is below: The solubility parameters can give an idea of whether or not a compound will be soluble inOnly a solvent or if one solvent is similar to another. If the values of the solute and the solvent have a difference around 5.1 MPa1/2, then the solute should be soluble in the solvent. Solvent and Cannabinoid Solubility Parameters When extracting cannabinoids, a process known as winterization can be used to precipitate out unwanted compounds. The waxes and fats from the plant material will precipitate as the material sits in a solvent in cold temperatures. According to Hildebrand solubility parameters, these compounds precipitate out since their solubility parameters are lower than that of certain solvents. They do not match the difference of 5.1 MPa1/2 so they come out of solution during winterization. The following table lists the solubility parameters of some common solvents, cannabinoids, and terpenes: Compound MPa1/2 Compound MPa1/2 Solvents a-pinene 16.16 n-Heptane 15.40 p-cymene 16.57 Ethanol 26.20 B-myrcene 21.46 710 Spirits 25.66 B-caryophyllene 21.41 Isopropyl Alcohol 23.6 d-Limonene 24.46 Acetone 19.9 Pulegone 25.65 Butane 14.1 1,8-cineole 18.96 Cannabinoids and Terpenes a-pinene 16.15 THC Information22.09 a-terpineol 17.10 CBD 24.34 terpineol-4-ol 18.21 THCA 24.14 p-cymene 24.20 CBDA 25.77 Flavonoids Waxes 16.36 apigenin 20.80 Chlorophyll 16.98 quercetin 25.89 Limonene 16.90 cannflavin A 10.35 ForLinalool 20.46 B-sitosterol 3.86 pg. -

Butane Propane
Safety Data Sheet Product: Butane-Propane Mixture Page: 1 of 6 Issued: 12-November-2015 SECTION 1 - IDENTIFICATION: PRODUCT IDENTIFIER AND CHEMICAL IDENTITY The Coleman Company, Inc. 3600 N Hydraulic Wichita, Kansas 67219 Phone: 1 800 835 3278 (bus hours) Chemical Nature: Petroleum Gases, Liquefied Trade Name: Butane-Propane Mixture Product Use: Disposable cartridges for use in portable gas appliances Creation Date: 13-March-2013 Revision Date: 12-November-2015 SECTION 2 - HAZARD IDENTIFICATION UN Number: 2037, Receptacles, small, containing gas (gas cartridges) flammable, without release device, not refillable and not exceeding 1 L capacity Classification: Flammable Gases, Category 1 SUSDP Classification: S6 Hazard DANGER Statements Extremely flammable gas. Harmful if inhaled. Causes skin irritation. Causes serious eye irritation. Precautionary Keep out of the reach of children. Statements Keep container in a well-ventilated place at a temperature not exceeding 48oC. Keep away from heat and ignition sources - No Smoking. In case of fire, use dry chemical, water fog or foam. Use only in well ventilated areas. Wash thoroughly with soap and water after handling and before eating, drinking or using tobacco. In case of insufficient ventilation, wear suitable respiratory equipment. Remove to fresh air immediately. Get medical attention immediately. Do NOT induce vomiting. Get immediate medical attention. If frostbite, call a physician. Hold eyelids apart and flush eyes with plenty of water for at least 15 minutes. Get medical attention. Avoid -

Safety Data Sheet
SAFETY DATA SHEET Flammable Gas Mixture: Acetone / Argon / C6+(Hexanes Plus) / Carbon Dioxide / Ethane / Ethanol / Helium / Hydrogen / Isobutane / Isopentane / Isopropyl Alcohol / Methane / N-Butane / N-Pentane / Neopentane / Nitrogen / Propane Section 1. Identification GHS product identifier : Flammable Gas Mixture: Acetone / Argon / C6+(Hexanes Plus) / Carbon Dioxide / Ethane / Ethanol / Helium / Hydrogen / Isobutane / Isopentane / Isopropyl Alcohol / Methane / N-Butane / N-Pentane / Neopentane / Nitrogen / Propane Other means of : Not available. identification Product use : Synthetic/Analytical chemistry. SDS # : 018560 Supplier's details : Airgas USA, LLC and its affiliates 259 North Radnor-Chester Road Suite 100 Radnor, PA 19087-5283 1-610-687-5253 24-hour telephone : 1-866-734-3438 Section 2. Hazards identification OSHA/HCS status : This material is considered hazardous by the OSHA Hazard Communication Standard (29 CFR 1910.1200). Classification of the : FLAMMABLE GASES - Category 1 substance or mixture GASES UNDER PRESSURE - Compressed gas TOXIC TO REPRODUCTION (Fertility) - Category 2 TOXIC TO REPRODUCTION (Unborn child) - Category 2 AQUATIC HAZARD (LONG-TERM) - Category 3 GHS label elements Hazard pictograms : Signal word : Danger Hazard statements : Extremely flammable gas. Contains gas under pressure; may explode if heated. May form explosive mixtures in Air. May displace oxygen and cause rapid suffocation. May increase respiration and heart rate. Suspected of damaging fertility or the unborn child. Harmful to aquatic life with long lasting effects. Precautionary statements General : Read and follow all Safety Data Sheets (SDS’S) before use. Read label before use. Keep out of reach of children. If medical advice is needed, have product container or label at hand. Close valve after each use and when empty. -

Section 5. Fire-Fighting Measures Extinguishing Media Suitable Extinguishing : Use an Extinguishing Agent Suitable for the Surrounding Fire
SAFETY DATA SHEET Flammable Gas Mixture: 1-Butene / Acetylene / Carbon Monoxide / Ethane / Ethylene / Hydrogen / Isobutane / Methane / Methyl Acetylene / N-Butane / Propadiene / Propane / Propylene Section 1. Identification GHS product identifier : Flammable Gas Mixture: 1-Butene / Acetylene / Carbon Monoxide / Ethane / Ethylene / Hydrogen / Isobutane / Methane / Methyl Acetylene / N-Butane / Propadiene / Propane / Propylene Other means of : Not available. identification Product use : Synthetic/Analytical chemistry. SDS # : 012433 Supplier's details : Airgas USA, LLC and its affiliates 259 North Radnor-Chester Road Suite 100 Radnor, PA 19087-5283 1-610-687-5253 Emergency telephone : 1-866-734-3438 number (with hours of operation) Section 2. Hazards identification OSHA/HCS status : This material is considered hazardous by the OSHA Hazard Communication Standard (29 CFR 1910.1200). Classification of the : FLAMMABLE GASES - Category 1 substance or mixture GASES UNDER PRESSURE - Compressed gas SPECIFIC TARGET ORGAN TOXICITY (SINGLE EXPOSURE) (Narcotic effects) - Category 3 GHS label elements Hazard pictograms : Signal word : Danger Hazard statements : Extremely flammable gas. Contains gas under pressure; may explode if heated. May displace oxygen and cause rapid suffocation. May cause drowsiness and dizziness. Precautionary statements General : Read and follow all Safety Data Sheets (SDS’S) before use. Read label before use. Keep out of reach of children. If medical advice is needed, have product container or label at hand. Close valve after each use and when empty. Use equipment rated for cylinder pressure. Do not open valve until connected to equipment prepared for use. Use a back flow preventative device in the piping. Use only equipment of compatible materials of construction. Approach suspected leak area with caution. -

Butane-Commercial Grade
SAFETY DATA SHEET 1. Identification Product identifier Butane (Commercial Grade) Other means of identification SDS number 5 Recommended use Fuel. Recommended restrictions None known. Manufacturer / Importer / Supplier / Distributor information Company name DCP Midstream Address 370 17 Street Suite 2500 Denver, CO 80202 Telephone (303) 595-3331 E-mail [email protected] Contact person Mark Prewitt Emergency phone number CHEMTREC - 24 HOURS: 800-424-9300 2. Hazard(s) identification Physical hazards Flammable gases Category 1 Gases under pressure Liquefied gas Health hazards Not classified. OSHA hazard(s) Simple asphyxiant Label elements Hazard symbol Signal word Danger Hazard statement Extremely flammable gas. Contains gas under pressure; may explode if heated. May displace oxygen and cause rapid suffocation. Precautionary statement Prevention Keep away from heat/sparks/open flames/hot surfaces. - No smoking. Response Leaking gas fire: Do not extinguish, unless leak can be stopped safely. Eliminate all ignition sources if safe to do so. Storage Store in a well-ventilated place. Disposal Dispose of contents/container in accordance with local/regional/national/international regulations. Hazard(s) not otherwise Not classified. classified (HNOC) 3. Composition/information on ingredients Mixture Hazardous components Chemical name Common name and synonyms CAS number % Butane 106-97-8 94 Isobutane 75-28-5 4 Pentane 109-66-1 2 Propane 74-98-6 1 Butane (Commercial Grade) 1/ 9 91653 Version#: 01 Revision date: - 10-19-2016 Issue Date: 11-28-2012 Composition comments All concentrations are in percent by weight unless ingredient is a gas. Gas concentrations are in percent by volume. 4. First-aid measures Inhalation Immediately remove from further exposure. -

The Detailed Kinetics of the Partial Oxidation Reactions: Benzene/Butane to Maleic Anhydride Ethylene to Ethylene Oxide
The Detailed Kinetics of the Partial Oxidation Reactions: Benzene/Butane to Maleic Anhydride Ethylene to Ethylene Oxide Ken Waugh Emeritus Professor of Physical Chemistry ICI Central Instruments Research Lab Bozedown House Berkshire The Oxidation of Benzene to Maleic Anhydride Over Vanadium Pentoxide D(V‐O) = 600 kJmol‐1 D A Dowden mechanism of benzene oxidation to MA using molecularly adsorbed O2 Maleic anhydride temperature programmed desorption spectra from V2O5: adsorption temperature (a) 135, (b) 150, (c) 160, (d) 170, (e) 180, (f) 190, (g) 200 C Benzene temperature programmed desorption spectra: curve (a) benzene, curve (b) maleic anhydride Desorption spectra following the adsorption of benzene at (a) 75, (b) 90 and (c) 100 C. Curves 1and 2 are benzene; curve (c) is maleic anhydride Activation Energy 31 kJmol‐1 The Oxidation of n‐butane to Maleic Anhydride • Anaerobic Selectivity 80% The (100) face of (VO)2P2O7 –the active face Recirculating Solids Reactor for n‐butane Oxidation to Maleic Anhydride ‐ DuPont Schematic Representation of the Mars and van Krevelen Mechanism Oxygen desorption from (VO)2P2O7 Anaerobic Temperature Programmed Oxidation of n‐ butane over the (VO)2P2O7 Catalyst Anaerobic Temperature Programmed Oxidation of but‐1‐ene over the (VO)2P2O7 Catalyst Anaerobic Temperature Programmed Oxidation of but‐1,3‐diene over the (VO)2P2O7 Catalyst Side View of n‐butane Adsorbed End‐on Identification of the Origins of Selectivity in Ethylene Epoxidation on Promoted and Unpromoted Ag/α‐Al2O3 Catalysts: a detailed kinetic, mechanistic -

TERNARY LIQUID-LIQUID EQUILIBRIA for the SYSTEMS of AQUEOUS METHANOL SOLUTIONS and PROPANE OR N-BUTANE
w = mass of solid per unit area measured from 3) Carslaw, H. S. and J. C. Jaeger: "Conduction of Heat in medium [g/cm2] Solids", 2nd ed , p. 285, Oxford Univ. Press (1959). Wi, w* = value of w at cake surface and total mass 4) Crank, J. : "The Mathematics of Diffusion", Oxford Univ. of solid per unit area [g/cm2] Press (1956). x = distance from medium [cm] 5) Fujita, H.: /. Chem. Phys., 21, 700 (1953). x% = thickness of cake [cm] 6) Hamill, T. D. and S. G. Bankoff: Ind. Eng. Chem. Funda- mentals, 3, 177 (1964). a = defined by Eq. (30) [-] 7) Lee,E. S. andL. T. Fan: Can. J. Chem. Eng.,46, 200(1968). /3 = 0\b\ [-] 8) Okamura, S. and M. Shirato: Kagaku Kogaku, 19, 104, 111 s = porosity [-] (1955). r] - defined by Eq. (14) [-] 9) Satoh, T. and K. Atsumi: J. Chem. Eng. Japan, 3, 92 (1970). 0 = non-dimensional time, defined by Eq. (14) [-] 10) Shirato, M. and T. Aragaki: Advanced Chemical Engineer- x = b/VT [-] ing (Japan), 8, 121 (1974). li = viscosity of filtrate [g/cm-sec] ll) Shirato, M. and T. Murase: ibid, 8, 153 (1974). P, ps = density of filtrate and of solid [g/cm3] 12) Shirato, M., M. Sambuichi, T. Aragaki and H. Kato: Kagaku Kogaku, 31, 359 (1967). 13) idem: AlChEJ., 15, 405 (1969). Literature Cited 14) Shirato, M. and M. Sambuichi: Kagaku Kogaku, 27, 470 1) Atsumi, K., T. Akiyama and M. Miyagawa: /. Chem. Eng. (1963). Japan, 6, 236 (1973). 15) Smiles, D. E.: Chem. Eng. ScL, 25, 985 (1970). -

SAFETY DATA SHEET Propane & Butane Gas Refill
Revision date: 5/24/2016 Revision: 1 SAFETY DATA SHEET Propane & Butane Gas Refill According to Appendix D, OSHA Hazard Communication Standard 29 CFR §1910.1200 1. Identification Product identifier Product name Propane & Butane Gas Refill Product number 71006402, ZP Recommended use of the chemical and restrictions on use Application Cleaning agent. Uses advised against No specific uses advised against are identified. Details of the supplier of the safety data sheet Supplier Allied Electronics, Inc. Allied Electronics, Inc. 7151 Jack Newell Blvd. S. Fort Worth, Texas 76118 U.S.A. www.alliedelec.com Tel: +1 (866) 433-5722 Emergency telephone number Emergency telephone +1 202 464 2554 (USA only) +44 1235 239670 2. Hazard(s) identification Classification of the substance or mixture Physical hazards Flam. Aerosol 1 - H222 Press. Gas, Compressed - H280 Health hazards Not Classified Environmental hazards Not Classified Label elements Pictogram Signal word Danger Hazard statements H222 Extremely flammable aerosol. H280 Contains gas under pressure; may explode if heated. Precautionary statements P210 Keep away from heat, sparks, open flames and hot surfaces. No smoking. P211 Do not spray on an open flame or other ignition source. P251 Pressurized container: Do not pierce or burn, even after use P410+P403 Protect from sunlight. Store in a well-ventilated place. P412 Do not expose to temperatures exceeding 50°C/122°F. P102 Keep out of reach of children. Other hazards This product does not contain any substances classified as PBT or vPvB. 1/12 Revision date: 5/24/2016 Revision: 1 Propane & Butane Gas Refill 3. Composition/information on ingredients Mixtures Petroleum gases, liquefied 60-100% CAS number: 68476-85-7 Classification Flam. -

Safety Alert 13-1
NEW JERSEY DEPARTMENT OF COMMUNITY AFFAIRS DIVISION OF FIRE SAFETY OFFICE OF THE STATE FIRE MARSHAL SSAAFFEETTYY AALLEERRTT PO Box 809, Trenton, NJ 08625-0809 (609) 633-6070 SAFETY ALERT 13-1 Butane Hash Oil Extraction Hazards Issued January, 2013 Over the last few months, law enforcement agencies have noted a marked increase in calls for service related to the use of an improvised hash oil preparation method known within the marijuana user community as “BHO” or Butane Honey Oil extraction. Honey oil is a concentrated substance derived from Cannabis (Marijuana). The texture varies from crystal (gloss) amber to gold resin (smoother). Hash oil is a resinous mixture of cannabinoids produced by a solvent extraction of Cannabis. Hash oil is a concentrated product with a high tetrahydrocannabinol (THC) content, the active ingredient in marijuana that produces the “high. Honey oil is a specific type of hash oil extracted with butane and is typically smoked. The process involves the use of an extractor tube, which can either be purchased commercially or homemade. The tube is typically made out of 1.75” diameter PVC pipe, steel pipe, glass or plastic and is usually about 1’ long. If PVC or metal pipe is used, end caps are also required. The finished appearance is similar to a pipe bomb. The extractor tube is filled with marijuana and a volatile solvent is injected into the top of the tube to extract a resinous mixture of cannabinoids. Individual solvents can include isopropyl alcohol, ethanol, methyl alcohol, and butane or isobutene. The resin collects at the bottom of the extractor which is usually lined with a coffee filter or other suitable screen, then removed and heated to evaporate the remaining solvent and “purify” the end product. -

SAFETY DATA SHEET Gosystem Butane-Propane Mix Gas Cartridge According to Regulation (EC) No 1907/2006, Annex II, As Amended by Regulation (EU) No 453/2010
Revision date: 04/06/2015 Revision: 1 Supersedes date: 01/12/2014 SAFETY DATA SHEET GoSystem Butane-Propane Mix Gas Cartridge According to Regulation (EC) No 1907/2006, Annex II, as amended by Regulation (EU) No 453/2010 SECTION 1: Identification of the substance/mixture and of the company/undertaking 1.1. Product identifier Product name GoSystem Butane-Propane Mix Gas Cartridge Chemical name Petroleum gases, liquefied Product number 2350 Synonyms; trade names Hydrocarbons C3-C4,LPG Container size 350g 1.2. Relevant identified uses of the substance or mixture and uses advised against Identified uses Fuel Uses advised against No specific uses advised against are identified. 1.3. Details of the supplier of the safety data sheet Supplier Go Gas Ltd. Unit 1 B, East Tame Business Park, Newton, Hyde, Cheshire, SK14 4GX UK. www.go-system.co.uk T: +44 (0) 161 367 1315 F: +44 (0) 161 367 1316 [email protected] 1.4. Emergency telephone number Emergency telephone +44 (0) 161 367 1315 Mon - Fri 09:00-17:00h (UK) SECTION 2: Hazards identification 2.1. Classification of the substance or mixture Classification Physical hazards Flam. Gas 1 - H220 Press. Gas, Liquefied - H280 Health hazards Not Classified Environmental hazards Not Classified Classification (67/548/EEC or F+; R12 1999/45/EC) 2.2. Label elements 1/10 Revision date: 04/06/2015 Revision: 1 Supersedes date: 01/12/2014 GoSystem Butane-Propane Mix Gas Cartridge Pictogram Signal word Danger Hazard statements H220 Extremely flammable gas. H280 Contains gas under pressure; may explode if heated. Exemptions from CLP Article The following are not required for labelling:,H280,- 1.3.2.