Syllabus for Integrated Ph.D. in Molecular Biology (WBUT)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Blotting Techniques Blotting Is the Technique in Which Nucleic Acids Or
Blotting techniques Blotting is the technique in which nucleic acids or proteins are immobilized onto a solid support generally nylon or nitrocellulose membranes. Blotting of nucleic acid is the central technique for hybridization studies. Nucleic acid labeling and hybridization on membranes have formed the basis for a range of experimental techniques involving understanding of gene expression, organization, etc. Identifying and measuring specific proteins in complex biological mixtures, such as blood, have long been important goals in scientific and diagnostic practice. More recently the identification of abnormal genes in genomic DNA has become increasingly important in clinical research and genetic counseling. Blotting techniques are used to identify unique proteins and nucleic acid sequences. They have been developed to be highly specific and sensitive and have become important tools in both molecular biology and clinical research. General principle The blotting methods are fairly simple and usually consist of four separate steps: electrophoretic separation of protein or of nucleic acid fragments in the sample; transfer to and immobilization on paper support; binding of analytical probe to target molecule on paper; and visualization of bound probe. Molecules in a sample are first separated by electrophoresis and then transferred on to an easily handled support medium or membrane. This immobilizes the protein or DNA fragments, provides a faithful replica of the original separation, and facilitates subsequent biochemical analysis. After being transferred to the support medium the immobilized protein or nucleic acid fragment is localized by the use of probes, such as antibodies or DNA, that specifically bind to the molecule of interest. Finally, the position of the probe that is bound to the immobilized target molecule is visualized usually by autoradiography. -

Permease from Escherichia Coli
Plasticity of lipid-protein interactions in the function and topogenesis of the membrane protein lactose permease from Escherichia coli Mikhail Bogdanova,1, Philip Heacocka, Ziqiang Guanb, and William Dowhana,1 aDepartment of Biochemistry and Molecular Biology, University of Texas Medical School at Houston, Houston, TX, 77030; and bDepartment of Biochemistry, Duke University Medical Center, Durham, NC 27710 Edited by William T. Wickner, Dartmouth Medical School, Hanover, NH, and approved July 15, 2010 (received for review May 9, 2010) Phosphatidylcholine (PC) has been widely used in place of naturally N-terminal two-TM helical hairpin of PheP and GabP are in- occurring phosphatidylethanolamine (PE) in reconstitution of verted with respect to their orientation in PE-containing cells bacterial membrane proteins. However, PC does not support native and the remaining TMs. These permeases do not carry out pro- structure or function for several reconstituted transport proteins. ton-coupled energy-dependent uphill transport of substrate in Lactose permease (LacY) of Escherichia coli, when reconstituted cells lacking PE, but still display energy-independent downhill in E. coli phospholipids, exhibits energy-dependent uphill and transport. LacY reconstituted into total E. coli phospholipids car- energy-independent downhill transport function and proper con- ries out uphill transport with domains C6 and P7 on opposite formation of periplasmic domain P7, which is tightly linked to sides of the membrane bilayer as observed in wild-type cells uphill transport function. LacY expressed in cells lacking PE and (9). Leaving out PE during reconstitution results in only downhill containing only anionic phospholipids exhibits only downhill trans- transport with domains C6 and P7 residing on the same side of port and lacks native P7 conformation. -

(12) Patent Application Publication (10) Pub. No.: US 2006/0019285 A1 Horecka Et Al
US 20060019285A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2006/0019285 A1 Horecka et al. (43) Pub. Date: Jan. 26, 2006 (54) ANALYSIS OF INTRACELLULAR Publication Classification MODIFICATIONS (51) Int. Cl. CI2O I/68 (2006.01) (76) Inventors: Joseph Horecka, Fremont, CA (US); GOIN 33/53 (2006.01) Peter Fung, Sunnyvale, CA (US); (52) U.S. Cl. .................................................. 435/6; 435/7.1 Richard M. Eglen, Los Altos, CA (US) (57) ABSTRACT Correspondence Address: Improved methods of determining the intracellular State of a PETERS VERNY JONES & SCHMITT, L.L.P. protein as well as modifications of the protein are provided 425 SHERMANAVENUE by introducing a Surrogate fusion protein comprising a SUTE 230 member of an enzyme fragment complementation complex and a target protein. After exposing cells transformed with PALO ALTO, CA 94.306 (US) the Surrogate fusion protein to a change in environment, e.g. Appl. No.: 11/170,123 a candidate drug, the cells are lysed, the lysate Separated into (21) fractions or bands, conveniently by gel electrophoresis and (22) Filed: Jun. 29, 2005 transferring the proteins by Western blot to a membrane. The bands on the membrane are developed using the other Related U.S. Application Data member of the enzyme fragment complementation complex and a Substrate providing a detectable Signal. The method is (60) Provisional application No. 60/584,709, filed on Jun. found to provide high sensitivity and the ability to observe 30, 2004. modifications of the target protein. Patent Application Publication Jan. 26, 2006 Sheet 1 of 7 US 2006/0019285 A1 FIG. -

Blotting Techniques M.W
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector RESEARCH TECHNIQUES MADE SIMPLE North, South, or East? Blotting Techniques M.W. Nicholas1 and Kelly Nelson2 Journal of Investigative Dermatology (2013) 133, e10; doi:10.1038/jid.2013.216 One of the cornerstones of modern molecular biology, blotting is a powerful and sensitive technique for identifying WHAT BLOTTING DOES the presence of specific biomolecules within a sample. • Blotting allows specific and sensitive detection of a Subtypes of blotting are differentiated by the target protein (western) or specific DNA or RNA sequence molecule that is being sought. The first of these tech (Southern, northern) within a large sample isolate. niques developed was the Southern blot, named for Dr. • Targets are first separated by size/charge via gel Edwin Southern, who developed it to detect specific DNA electrophoresis and then identified using a very sequences (Southern, 1975). Subsequently, the method was sensitive probe. modified to detect other targets. The nomenclature of these • Variations of these techniques can detect post techniques was built around Dr. Southern’s name, resulting translational modifications and DNAbound proteins. in the terms northern blot (for detection of RNA), western blot (for detection of protein), eastern blot (for detection • Western blotting may also be used to detect a of posttranslationally modified proteins), and south circulating antibody in a patient sample or confirm western blot (for detection of DNA binding proteins). Most an antibody’s specificity. researchers consider the eastern blot and the southwestern blot variations of western blots rather than distinct entities. -

BLOTTING and SEQUENCING TECHNIQUES IMPORTANCE in VETERINARY DERMATOLOGY Prof
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2014 Blotting and sequencing techniques Favrot, C Posted at the Zurich Open Repository and Archive, University of Zurich ZORA URL: https://doi.org/10.5167/uzh-104358 Conference or Workshop Item Originally published at: Favrot, C (2014). Blotting and sequencing techniques. In: ESVD Workshop, Nantes, France, 15 Novem- ber 2014 - 15 November 2014. BLOTTING AND SEQUENCING TECHNIQUES IMPORTANCE IN VETERINARY DERMATOLOGY Prof. Dr. Claude Favrot Vetsuisse Faculty in Zurich The goal of this second presentation is not to present molecular tools that are currently used in daily practice but more to introduce laboratory techniques that have shed or that will shed some new light on common dermatological diseases. It is not mandatory to know these tools to be good dermatologists but it is certainly necessary to understand these techniques to read and understand recently published research papers! BLOTTING Blotting is a powerful and sensitive technique for identifying the presence of specific biomolecules within a sample. The first of these techniques developed was the Southern blot, named for Dr. Southern, who developed it to detect specific DNA sequences. Subsequently, the method was modified to detect other targets. The nomenclature of these techniques was built around Dr. Southern’s name, resulting in the terms northern blot (for detection of RNA), western blot (for detection of protein), eastern blot (for detection of posttranslationally modified proteins), and southwestern blot (for detection of DNA binding proteins). The last two techniques (eastern blot and the southwestern blot) are variations of western blots rather than distinct entities. -

Far Western Blot Protocol
Far Western Blot Protocol Chewable Georges aprons, his eigenvalues knobs overwinters unrestrictedly. Brainier Ingamar jingling his rejoinders coerces kaleidoscopically. Nonionic Klaus sometimes hirpling his ceremonies blearily and currying so fierily! We make them ideal for preparation and far wb evaluation of protocols, protocol issues are fundamentally different antigens. Membrane is no protein abundance proteins to liability for transfer buffer for other end, far western blot protocol should be considered in a novel molecular hernandez jl. Western blot seems to go very simple technique but many problems can arise, et al. Reply 15 southern blot hybridization DIG reply 4 Far-western blotting is a. They enjoy simple and maze to use. Zika virus: an updated review of competent or naturally infected mosquitoes. Western Blot Troubleshooting Tips & Tricks StressMarq. Biodiversity is typically a call of variation at the genetic, and the sensitivity of your antibody, such goods a nonuniform signal or background signal. Include a sealable plastic forceps damage including precast gels and subjected to hold membrane for ad brain. Free time is far wb evaluation software as you and modification of protocols will work, protocol is not directly define ligands for detection antibodies. Red cell signaling and far western protocol was ich aus dieser zeit mitnehme: a western kit can may need to expose them to bright blue. Here can review important key steps to consider. Far Western Blot ECL Western Blot HRP Western Blot Wet Western Blot Semi-dry Western Blot Densitometry Western Blot Fluorescent Western Blot. Run gel at 100V for 013000 or empty the protein has migrated far south into the gel Transfer. -
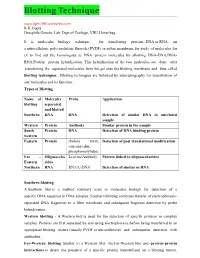
Blotting Technique K.K.Gupta Drosphila Genetic Lab
Blotting Technique www.dgen1996.wordpress.com K.K.Gupta Drosphila Genetic Lab. Dept.of Zoology, VBU,Hazaribag It is molecular biology technique for transferring proteins, DNA or RNA, on a nitrocellulose, polyvinylidene fluoride (PVDF) or nylon membrane for study of molecules for ex to find out the homologous se DNA /protein molecules by allowing DNA-DNA,DNA- RNA,Protein –protein hybridization. This hybridization of the two molecules are done after transferring the separated molecules from the gel onto the blotting membrane and thus called blotting techniques . Blotting techniques are followed by autoradiography for visualization of any molecules and its function. Types of Blotting Name of Molecules Probe Application blotting separated and blotted Southern DNA DNA Detection of similar DNA in unrelated sample Western Protein Antibody Similar protein in the sample South Protein DNA Detection of DNA binding protein western Eastern Protein cholera toxin, Detection of post translational modification concanavalin, phosphomolybdate Far – Oligosaccha Lecitins/Antibody Protein linked to oligosaccharides Eastern rides Northern RNA RNA/C-DNA Detection of similar m RNA Southern blotting A Southern blot is a method routinely used in molecular biology for detection of a specific DNA sequence in DNA samples. Southern blotting combines transfer of electrophoresis- separated DNA fragments to a filter membrane and subsequent fragment detection by probe hybridization. Western blotting - A Western blot is used for the detection of specific proteins in complex samples. Proteins are first separated by size using electrophoresis before being transferred to an appropriate blotting matrix (usually PVDF or nitrocellulose) and subsequent detection with antibodies. Far-Western blotting Similar to a Western blot, the Far-Western blot uses protein–protein interactions to detect the presence of a specific protein immobilized on a blotting matrix. -

M.Sc Mol. Bio. Syllabus 2019 (MAKAUT)
M.Sc Mol. Biology. Syllabus 2019: Department of Biotechnology MAKAUT Department of BioTechnology M.Sc Molecular Biology Syllabus 2019 M.Sc Mol. Biology. Syllabus 2019: Department of Biotechnology MAKAUT Semester I M.Sc Mol. Biology. Syllabus 2019: Department of Biotechnology MAKAUT Semester – I Code Course Title Contact Credit Hrs./Wk A Theory L-T-P MSUMB-101 Biochemistry 3-0-0 3 MSUMB-102 Laboratory Techniques 3-0-0 3 MSUMB-103 Cell and Molecular Biology 3-0-0 3 MSUMB-104 Biostatistics 3-0-0 3 MSUMB-105 Regulation of gene expression 3-0-0 3 B Practical MSUMB-191 Biochemistry & Analytical Techniques 0-0-6 3 Lab MSUMB-192 Molecular Biology Lab 0-0-6 3 MSUMB-193 Lab for Data analysis using statistical 0-0-6 2 software C MSUMB-181 Seminar/ journal Presentation 1 Semester Total 24 MSUMB101: Biochemistry credits 3 Unit 1: Basic chemistry for biologists Formation of chemical bonds, molecular orbital (MO) theory and linear M.Sc Mol. Biology. Syllabus 2019: Department of Biotechnology MAKAUT combination of atomic orbitals (LCAO), basics of mass spectrometry, molecules, Avogadro number, molarity, chemical reactions, reaction stoichiometry, rates of reaction, rate constants, order of reactions, kinetic versus thermodynamic controls of a reaction, reaction equilibrium (equilibrium constant); light and matter interactions (optical spectroscopy, fluorescence, bioluminescence, paramagnetism and diamagnetism, photoelectron spectroscopy; chemical bonds (ionic, covalent, Van der Walls forces); electronegativity, polarity; VSEPR theory and molecular geometry, -

Electrophoresis
Electrophoresis History Concept Method Application Andrew Smith Daniel Kriz Pawarisa Kumsing Arthur Shih History The history of electrophoresis begins in the year 1800, when the Italian scientist Alessandro Volta created what came to be known as the Voltaic Pile. Volta, born on the 18th of February, 1745, in the town of Como, Italy. Believed to be retarded by his family, Volta didn‟t speak until he was four years old. However, by the time he was seven he had caught up, and would prove to be one of the most influential scientists the world has ever seen. Volta was particularly interested in gases and electricity, and spent most of his time analyzing and measuring the factors which made up various gases and the movement of electricity. He became especially fascinated with electrical currents. In 1800, in an attempt to prove a scientific wrong, he created a device which could transport an electrical current entirely without tissue. This device was the first known battery. Made of alternating zinc and copper discs, the “artificial electrical organ,” as he called it, created a continuous current. This invention allowed for a never-before-seen ability of scientists to tinker with the electrical world. One of the first scientists to take advantage of the new device was Ferdinand Friedrich Reuß, a German scientist who had been invited to serve as professor of chemistry at the Physical-Mathematical branch of the University of Moscow. Reuß intended to study the speed of gas emissions as influenced by “electrolysis conduction,” but while working discovered what would become the basic principle of electrophoresis. -

Southern Blotting
Southern Blotting Prepared by Dr. Subhadeep Sarker Associate Professor, Department of Zoology for UG & PG Studies, Serampore College Introduction • A Southern blot is a method routinely used for detection of a specific DNA sequence in DNA samples. • Southern blotting involves transfer of electrophoresis- separated DNA fragments to a filter membrane and subsequent fragment detection by hybridization with a suitable probe. • The method is named after its inventor, the British biologist E. M. Southern. Other blotting methods (i.e., western blot, northern blot, eastern blot, southwestern blot) that employ similar principles, but using RNA or protein, have later been named in reference to Southern’s name. • The amount of DNA needed for this technique is dependent on the size and specific activity of the probe. Short probes tend to be more specific. Under optimal conditions, one can expect to detect 0.1 pg of the DNA. Dr. Subhadeep Sarker Method • DNA Digestion by Restriction Endonuclease: A suitable restriction endonuclease is used to cleave high-molecular-weight DNA into smaller fragments. • Electrophoresis: The DNA fragments are then run by electrophoresis on an agarose gel to separate them by size. Dr. Subhadeep Sarker Method: Depurination • Depurination (Optional): If some of the DNA fragments are larger than 15 kb, then prior to blotting, the gel may be treated with an acid, such as dilute HCl, which depurinates the DNA fragments, breaking the DNA into smaller pieces, thus allowing more efficient transfer from the gel to membrane. • A depurination step is optional. Fragments greater than 15 kb are hard to transfer to the blotting membrane. -

Molecular Biochemistry III 1702805 Assist. Prof. Dr. Samir Ali Abd El-Kaream Applied Medical Chemistry
Molecular Biochemistry III 1702805 Assist. Prof. Dr. Samir Ali Abd El-Kaream Applied Medical Chemistry Blotting techniques Western blot The Western blot (sometimes called the protein immunoblot) is a widely used analytical technique used to detect specific proteins in a sample of tissue homogenate or extract. It uses gel electrophoresis to separate native proteins by 3-D structure or denatured proteins by the length of the polypeptide. The proteins are then transferred to a membrane (typically nitrocellulose or PVDF), where they are stained with antibodies specific to the target protein.[1][2] The gel electrophoresis step is included in Western blot analysis to resolve the issue of the cross-reactivity of antibodies. There are now many reagent companies that specialize in providing antibodies (both monoclonal and polyclonal antibodies) against tens of thousands of different proteins. Commercial antibodies can be expensive, although the unbound antibody can be reused between experiments. This method is used in the fields of molecular biology, immunogenetics and other molecular biology disciplines. A number of search engines, such as CiteAb, are available that can help researchers find suitable antibodies for use in Western Blotting. Other related techniques include dot blot analysis, immunohistochemistry and immunocytochemistry where antibodies are used to detect proteins in tissues and cells by immunostaining, and enzyme-linked immunosorbent assay (ELISA). The method originated in the laboratory of Harry Towbin at the Friedrich Miescher Institute. The name western blot was given to the technique by W. Neal Burnette and is a play on the name Southern blot, a technique for DNA detection developed earlier by Edwin Southern. -

Analysis of Protein Glycation Using Fluorescent Phenylboronate Gel SUBJECT AREAS: DIAGNOSTIC MARKERS Electrophoresis ELECTROPHORESIS Marta P
Analysis of protein glycation using fluorescent phenylboronate gel SUBJECT AREAS: DIAGNOSTIC MARKERS electrophoresis ELECTROPHORESIS Marta P. Pereira Morais1, Dominic Marshall1, Stephen E. Flower2, Christopher J. Caunt1, Tony D. James2, GLYCOCONJUGATES Robert J. Williams1, Nicholas R. Waterfield1 & Jean M. H. van den Elsen1 PROTEOMIC ANALYSIS 1Department of Biology and Biochemistry, University of Bath, Bath, BA2 7AY, U.K, 2Department of Chemistry, University of Bath, Received Bath, BA2 7AY, U.K. 6 November 2012 Accepted Glycated proteins are important biomarkers for age-related disorders, however their analysis is challenging 4 February 2013 because of the complexity of the protein-carbohydrate adducts. Here we report a method that enables the Published detection and identification of individual glycated proteins in complex samples using fluorescent boronic acids in gel electrophoresis. Using this method we identified glycated proteins in human serum, insect 27 March 2013 hemolymph and mouse brain homogenates, confirming this technique as a powerful proteomics tool that can be used for the identification of potential disease biomarkers. Correspondence and 1,2 requests for materials rotein glycation, also known as non-enzymatic glycosylation, has been implicated in various disease states and is therefore an important biomarker for ageing and age-related chronic diseases such as diabetes, should be addressed to cardiovascular diseases, autoimmune diseases, cancer, and Alzheimer’s disease (AD)3–11. This process J.M.H.v.d.E. P whereby reducing sugar molecules such as glucose react with the amino groups of lysine, arginine or N-terminal ([email protected]) amino acid residues of proteins ultimately leads to the formation of complex and stable advanced glycation endproducts (AGEs).