ANOVA Assumptions
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
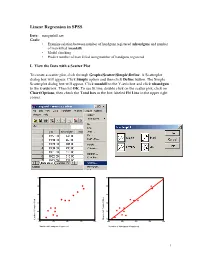
Linear Regression in SPSS
Linear Regression in SPSS Data: mangunkill.sav Goals: • Examine relation between number of handguns registered (nhandgun) and number of man killed (mankill) • Model checking • Predict number of man killed using number of handguns registered I. View the Data with a Scatter Plot To create a scatter plot, click through Graphs\Scatter\Simple\Define. A Scatterplot dialog box will appear. Click Simple option and then click Define button. The Simple Scatterplot dialog box will appear. Click mankill to the Y-axis box and click nhandgun to the x-axis box. Then hit OK. To see fit line, double click on the scatter plot, click on Chart\Options, then check the Total box in the box labeled Fit Line in the upper right corner. 60 60 50 50 40 40 30 30 20 20 10 10 Killed of People Number Number of People Killed 400 500 600 700 800 400 500 600 700 800 Number of Handguns Registered Number of Handguns Registered 1 Click the target button on the left end of the tool bar, the mouse pointer will change shape. Move the target pointer to the data point and click the left mouse button, the case number of the data point will appear on the chart. This will help you to identify the data point in your data sheet. II. Regression Analysis To perform the regression, click on Analyze\Regression\Linear. Place nhandgun in the Dependent box and place mankill in the Independent box. To obtain the 95% confidence interval for the slope, click on the Statistics button at the bottom and then put a check in the box for Confidence Intervals. -

Assessing Heterogeneity in Meta-Analysis: Q Statistic Or I2 Index? Tania Huedo-Medina University of Connecticut, [email protected]
University of Connecticut OpenCommons@UConn Center for Health, Intervention, and Prevention CHIP Documents (CHIP) 6-1-2006 Assessing heterogeneity in meta-analysis: Q statistic or I2 index? Tania Huedo-Medina University of Connecticut, [email protected] Julio Sanchez-Meca University of Murcia, Spane Fulgencio Marin-Martinez University of Murcia, Spain Juan Botella Autonoma University of Madrid, Spain Follow this and additional works at: https://opencommons.uconn.edu/chip_docs Part of the Psychology Commons Recommended Citation Huedo-Medina, Tania; Sanchez-Meca, Julio; Marin-Martinez, Fulgencio; and Botella, Juan, "Assessing heterogeneity in meta-analysis: Q statistic or I2 index?" (2006). CHIP Documents. 19. https://opencommons.uconn.edu/chip_docs/19 ASSESSING HETEROGENEITY IN META -ANALYSIS: Q STATISTIC OR I2 INDEX? Tania B. Huedo -Medina, 1 Julio Sánchez -Meca, 1 Fulgencio Marín -Martínez, 1 and Juan Botella 2 Running head: Assessing heterogeneity in meta -analysis 2006 1 University of Murcia, Spa in 2 Autónoma University of Madrid, Spain Address for correspondence: Tania B. Huedo -Medina Dept. of Basic Psychology & Methodology, Faculty of Psychology, Espinardo Campus, Murcia, Spain Phone: + 34 968 364279 Fax: + 34 968 364115 E-mail: [email protected] * This work has been supported by Plan Nacional de Investigaci ón Cient ífica, Desarrollo e Innovaci ón Tecnol ógica 2004 -07 from the Ministerio de Educaci ón y Ciencia and by funds from the Fondo Europeo de Desarrollo Regional, FEDER (Proyect Number: S EJ2004 -07278/PSIC). Assessing heterogeneity in meta -analysis 2 ASSESSING HETEROGENEITY IN META -ANALYSIS: Q STATISTIC OR I2 INDEX? Abstract In meta -analysis, the usual way of assessing whether a set of single studies are homogeneous is by means of the Q test. -

Auditor: an R Package for Model-Agnostic Visual Validation and Diagnostics
auditor: an R Package for Model-Agnostic Visual Validation and Diagnostics APREPRINT Alicja Gosiewska Przemysław Biecek Faculty of Mathematics and Information Science Faculty of Mathematics and Information Science Warsaw University of Technology Warsaw University of Technology Poland Faculty of Mathematics, Informatics and Mechanics [email protected] University of Warsaw Poland [email protected] May 27, 2020 ABSTRACT Machine learning models have spread to almost every area of life. They are successfully applied in biology, medicine, finance, physics, and other fields. With modern software it is easy to train even a complex model that fits the training data and results in high accuracy on test set. The problem arises when models fail confronted with the real-world data. This paper describes methodology and tools for model-agnostic audit. Introduced tech- niques facilitate assessing and comparing the goodness of fit and performance of models. In addition, they may be used for analysis of the similarity of residuals and for identification of outliers and influential observations. The examination is carried out by diagnostic scores and visual verification. Presented methods were implemented in the auditor package for R. Due to flexible and con- sistent grammar, it is simple to validate models of any classes. K eywords machine learning, R, diagnostics, visualization, modeling 1 Introduction Predictive modeling is a process that uses mathematical and computational methods to forecast outcomes. arXiv:1809.07763v4 [stat.CO] 26 May 2020 Lots of algorithms in this area have been developed and are still being develop. Therefore, there are countless possible models to choose from and a lot of ways to train a new new complex model. -

Statistical Analysis in JASP
Copyright © 2018 by Mark A Goss-Sampson. All rights reserved. This book or any portion thereof may not be reproduced or used in any manner whatsoever without the express written permission of the author except for the purposes of research, education or private study. CONTENTS PREFACE .................................................................................................................................................. 1 USING THE JASP INTERFACE .................................................................................................................... 2 DESCRIPTIVE STATISTICS ......................................................................................................................... 8 EXPLORING DATA INTEGRITY ................................................................................................................ 15 ONE SAMPLE T-TEST ............................................................................................................................. 22 BINOMIAL TEST ..................................................................................................................................... 25 MULTINOMIAL TEST .............................................................................................................................. 28 CHI-SQUARE ‘GOODNESS-OF-FIT’ TEST............................................................................................. 30 MULTINOMIAL AND Χ2 ‘GOODNESS-OF-FIT’ TEST. .......................................................................... -

A Robust Rescaled Moment Test for Normality in Regression
Journal of Mathematics and Statistics 5 (1): 54-62, 2009 ISSN 1549-3644 © 2009 Science Publications A Robust Rescaled Moment Test for Normality in Regression 1Md.Sohel Rana, 1Habshah Midi and 2A.H.M. Rahmatullah Imon 1Laboratory of Applied and Computational Statistics, Institute for Mathematical Research, University Putra Malaysia, 43400 Serdang, Selangor, Malaysia 2Department of Mathematical Sciences, Ball State University, Muncie, IN 47306, USA Abstract: Problem statement: Most of the statistical procedures heavily depend on normality assumption of observations. In regression, we assumed that the random disturbances were normally distributed. Since the disturbances were unobserved, normality tests were done on regression residuals. But it is now evident that normality tests on residuals suffer from superimposed normality and often possess very poor power. Approach: This study showed that normality tests suffer huge set back in the presence of outliers. We proposed a new robust omnibus test based on rescaled moments and coefficients of skewness and kurtosis of residuals that we call robust rescaled moment test. Results: Numerical examples and Monte Carlo simulations showed that this proposed test performs better than the existing tests for normality in the presence of outliers. Conclusion/Recommendation: We recommend using our proposed omnibus test instead of the existing tests for checking the normality of the regression residuals. Key words: Regression residuals, outlier, rescaled moments, skewness, kurtosis, jarque-bera test, robust rescaled moment test INTRODUCTION out that the powers of t and F tests are extremely sensitive to the hypothesized error distribution and may In regression analysis, it is a common practice over deteriorate very rapidly as the error distribution the years to use the Ordinary Least Squares (OLS) becomes long-tailed. -

ROC Curve Analysis and Medical Decision Making
ROC curve analysis and medical decision making: what’s the evidence that matters for evidence-based diagnosis ? Piergiorgio Duca Biometria e Statistica Medica – Dipartimento di Scienze Cliniche Luigi Sacco – Università degli Studi – Via GB Grassi 74 – 20157 MILANO (ITALY) [email protected] 1) The ROC (Receiver Operating Characteristic) curve and the Area Under the Curve (AUC) The ROC curve is the statistical tool used to analyse the accuracy of a diagnostic test with multiple cut-off points. The test could be based on a continuous diagnostic indicant, such as a serum enzyme level, or just on an ordinal one, such as a classification based on radiological imaging. The ROC curve is based on the probability density distributions, in actually diseased and non diseased patients, of the diagnostician’s confidence in a positive diagnosis, and upon a set of cut-off points to separate “positive” and “negative” test results (Egan, 1968; Bamber, 1975; Swets, Pickett, 1982; Metz, 1986; Zhou et al, 2002). The Sensitivity (SE) – the proportion of diseased turned out to be test positive – and the Specificity (SP) – the proportion of non diseased turned out to be test negative – will depend on the particular confidence threshold the observer applies to partition the continuously distributed perceptions of evidence into positive and negative test results. The ROC curve is the plot of all the pairs of True Positive Rates (TPR = SE), as ordinate, and False Positive Rates (FPR = (1 – SP)), as abscissa, related to all the possible cut-off points. An ROC curve represents all of the compromises between SE and SP can be achieved, changing the confidence threshold. -

Estimating the Variance of a Propensity Score Matching Estimator for the Average Treatment Effect
Observational Studies 4 (2018) 71-96 Submitted 5/15; Published 3/18 Estimating the variance of a propensity score matching estimator for the average treatment effect Ronnie Pingel [email protected] Department of Statistics Uppsala University Uppsala, Sweden Abstract This study considers variance estimation when estimating the asymptotic variance of a propensity score matching estimator for the average treatment effect. We investigate the role of smoothing parameters in a variance estimator based on matching. We also study the properties of estimators using local linear estimation. Simulations demonstrate that large gains can be made in terms of mean squared error, bias and coverage rate by prop- erly selecting smoothing parameters. Alternatively, a residual-based local linear estimator could be used as an estimator of the asymptotic variance. The variance estimators are implemented in analysis to evaluate the effect of right heart catheterisation. Keywords: Average Causal Effect, Causal Inference, Kernel estimator 1. Introduction Matching estimators belong to a class of estimators of average treatment effects (ATEs) in observational studies that seek to balance the distributions of covariates in a treatment and control group (Stuart, 2010). In this study we consider one frequently used matching method, the simple nearest-neighbour matching with replacement (Rubin, 1973; Abadie and Imbens, 2006). Rosenbaum and Rubin (1983) show that instead of matching on covariates directly to remove confounding, it is sufficient to match on the propensity score. In observational studies the propensity score must almost always be estimated. An important contribution is therefore Abadie and Imbens (2016), who derive the large sample distribution of a nearest- neighbour propensity score matching estimator when using the estimated propensity score. -

Testing Normality: a GMM Approach Christian Bontempsa and Nour
Testing Normality: A GMM Approach Christian Bontempsa and Nour Meddahib∗ a LEEA-CENA, 7 avenue Edouard Belin, 31055 Toulouse Cedex, France. b D´epartement de sciences ´economiques, CIRANO, CIREQ, Universit´ede Montr´eal, C.P. 6128, succursale Centre-ville, Montr´eal (Qu´ebec), H3C 3J7, Canada. First version: March 2001 This version: June 2003 Abstract In this paper, we consider testing marginal normal distributional assumptions. More precisely, we propose tests based on moment conditions implied by normality. These moment conditions are known as the Stein (1972) equations. They coincide with the first class of moment conditions derived by Hansen and Scheinkman (1995) when the random variable of interest is a scalar diffusion. Among other examples, Stein equation implies that the mean of Hermite polynomials is zero. The GMM approach we adopt is well suited for two reasons. It allows us to study in detail the parameter uncertainty problem, i.e., when the tests depend on unknown parameters that have to be estimated. In particular, we characterize the moment conditions that are robust against parameter uncertainty and show that Hermite polynomials are special examples. This is the main contribution of the paper. The second reason for using GMM is that our tests are also valid for time series. In this case, we adopt a Heteroskedastic-Autocorrelation-Consistent approach to estimate the weighting matrix when the dependence of the data is unspecified. We also make a theoretical comparison of our tests with Jarque and Bera (1980) and OPG regression tests of Davidson and MacKinnon (1993). Finite sample properties of our tests are derived through a comprehensive Monte Carlo study. -

An Introduction to Logistic Regression: from Basic Concepts to Interpretation with Particular Attention to Nursing Domain
J Korean Acad Nurs Vol.43 No.2, 154 -164 J Korean Acad Nurs Vol.43 No.2 April 2013 http://dx.doi.org/10.4040/jkan.2013.43.2.154 An Introduction to Logistic Regression: From Basic Concepts to Interpretation with Particular Attention to Nursing Domain Park, Hyeoun-Ae College of Nursing and System Biomedical Informatics National Core Research Center, Seoul National University, Seoul, Korea Purpose: The purpose of this article is twofold: 1) introducing logistic regression (LR), a multivariable method for modeling the relationship between multiple independent variables and a categorical dependent variable, and 2) examining use and reporting of LR in the nursing literature. Methods: Text books on LR and research articles employing LR as main statistical analysis were reviewed. Twenty-three articles published between 2010 and 2011 in the Journal of Korean Academy of Nursing were analyzed for proper use and reporting of LR models. Results: Logistic regression from basic concepts such as odds, odds ratio, logit transformation and logistic curve, assumption, fitting, reporting and interpreting to cautions were presented. Substantial short- comings were found in both use of LR and reporting of results. For many studies, sample size was not sufficiently large to call into question the accuracy of the regression model. Additionally, only one study reported validation analysis. Conclusion: Nurs- ing researchers need to pay greater attention to guidelines concerning the use and reporting of LR models. Key words: Logit function, Maximum likelihood estimation, Odds, Odds ratio, Wald test INTRODUCTION The model serves two purposes: (1) it can predict the value of the depen- dent variable for new values of the independent variables, and (2) it can Multivariable methods of statistical analysis commonly appear in help describe the relative contribution of each independent variable to general health science literature (Bagley, White, & Golomb, 2001). -

Njit-Etd2007-041
Copyright Warning & Restrictions The copyright law of the United States (Title 17, United States Code) governs the making of photocopies or other reproductions of copyrighted material. Under certain conditions specified in the law, libraries and archives are authorized to furnish a photocopy or other reproduction. One of these specified conditions is that the photocopy or reproduction is not to be “used for any purpose other than private study, scholarship, or research.” If a, user makes a request for, or later uses, a photocopy or reproduction for purposes in excess of “fair use” that user may be liable for copyright infringement, This institution reserves the right to refuse to accept a copying order if, in its judgment, fulfillment of the order would involve violation of copyright law. Please Note: The author retains the copyright while the New Jersey Institute of Technology reserves the right to distribute this thesis or dissertation Printing note: If you do not wish to print this page, then select “Pages from: first page # to: last page #” on the print dialog screen The Van Houten library has removed some of the personal information and all signatures from the approval page and biographical sketches of theses and dissertations in order to protect the identity of NJIT graduates and faculty. ABSTRACT PROBLEMS RELATED TO EFFICACY MEASUREMENT AND ANALYSES by Sibabrata Banerjee In clinical research it is very common to compare two treatments on the basis of an efficacy vrbl Mr pfll f Χ nd Υ dnt th rpn f ptnt n th t trtnt A nd rptvl th -

Power Comparisons of Shapiro-Wilk, Kolmogorov-Smirnov, Lilliefors and Anderson-Darling Tests
Journal ofStatistical Modeling and Analytics Vol.2 No.I, 21-33, 2011 Power comparisons of Shapiro-Wilk, Kolmogorov-Smirnov, Lilliefors and Anderson-Darling tests Nornadiah Mohd Razali1 Yap Bee Wah 1 1Faculty ofComputer and Mathematica/ Sciences, Universiti Teknologi MARA, 40450 Shah Alam, Selangor, Malaysia E-mail: nornadiah@tmsk. uitm. edu.my, yapbeewah@salam. uitm. edu.my ABSTRACT The importance of normal distribution is undeniable since it is an underlying assumption of many statistical procedures such as I-tests, linear regression analysis, discriminant analysis and Analysis of Variance (ANOVA). When the normality assumption is violated, interpretation and inferences may not be reliable or valid. The three common procedures in assessing whether a random sample of independent observations of size n come from a population with a normal distribution are: graphical methods (histograms, boxplots, Q-Q-plots), numerical methods (skewness and kurtosis indices) and formal normality tests. This paper* compares the power offour formal tests of normality: Shapiro-Wilk (SW) test, Kolmogorov-Smirnov (KS) test, Lillie/ors (LF) test and Anderson-Darling (AD) test. Power comparisons of these four tests were obtained via Monte Carlo simulation of sample data generated from alternative distributions that follow symmetric and asymmetric distributions. Ten thousand samples ofvarious sample size were generated from each of the given alternative symmetric and asymmetric distributions. The power of each test was then obtained by comparing the test of normality statistics with the respective critical values. Results show that Shapiro-Wilk test is the most powerful normality test, followed by Anderson-Darling test, Lillie/ors test and Kolmogorov-Smirnov test. However, the power ofall four tests is still low for small sample size. -

A Quantitative Validation Method of Kriging Metamodel for Injection Mechanism Based on Bayesian Statistical Inference
metals Article A Quantitative Validation Method of Kriging Metamodel for Injection Mechanism Based on Bayesian Statistical Inference Dongdong You 1,2,3,* , Xiaocheng Shen 1,3, Yanghui Zhu 1,3, Jianxin Deng 2,* and Fenglei Li 1,3 1 National Engineering Research Center of Near-Net-Shape Forming for Metallic Materials, South China University of Technology, Guangzhou 510640, China; [email protected] (X.S.); [email protected] (Y.Z.); fl[email protected] (F.L.) 2 Guangxi Key Lab of Manufacturing System and Advanced Manufacturing Technology, Guangxi University, Nanning 530003, China 3 Guangdong Key Laboratory for Advanced Metallic Materials processing, South China University of Technology, Guangzhou 510640, China * Correspondence: [email protected] (D.Y.); [email protected] (J.D.); Tel.: +86-20-8711-2933 (D.Y.); +86-137-0788-9023 (J.D.) Received: 2 April 2019; Accepted: 24 April 2019; Published: 27 April 2019 Abstract: A Bayesian framework-based approach is proposed for the quantitative validation and calibration of the kriging metamodel established by simulation and experimental training samples of the injection mechanism in squeeze casting. The temperature data uncertainty and non-normal distribution are considered in the approach. The normality of the sample data is tested by the Anderson–Darling method. The test results show that the original difference data require transformation for Bayesian testing due to the non-normal distribution. The Box–Cox method is employed for the non-normal transformation. The hypothesis test results of the calibrated kriging model are more reliable after data transformation. The reliability of the kriging metamodel is quantitatively assessed by the calculated Bayes factor and confidence.