Approximation-Based Similarity Search for 3-D Surface Segments1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Chapter 5 Dimensional Analysis and Similarity
Chapter 5 Dimensional Analysis and Similarity Motivation. In this chapter we discuss the planning, presentation, and interpretation of experimental data. We shall try to convince you that such data are best presented in dimensionless form. Experiments which might result in tables of output, or even mul- tiple volumes of tables, might be reduced to a single set of curves—or even a single curve—when suitably nondimensionalized. The technique for doing this is dimensional analysis. Chapter 3 presented gross control-volume balances of mass, momentum, and en- ergy which led to estimates of global parameters: mass flow, force, torque, total heat transfer. Chapter 4 presented infinitesimal balances which led to the basic partial dif- ferential equations of fluid flow and some particular solutions. These two chapters cov- ered analytical techniques, which are limited to fairly simple geometries and well- defined boundary conditions. Probably one-third of fluid-flow problems can be attacked in this analytical or theoretical manner. The other two-thirds of all fluid problems are too complex, both geometrically and physically, to be solved analytically. They must be tested by experiment. Their behav- ior is reported as experimental data. Such data are much more useful if they are ex- pressed in compact, economic form. Graphs are especially useful, since tabulated data cannot be absorbed, nor can the trends and rates of change be observed, by most en- gineering eyes. These are the motivations for dimensional analysis. The technique is traditional in fluid mechanics and is useful in all engineering and physical sciences, with notable uses also seen in the biological and social sciences. -
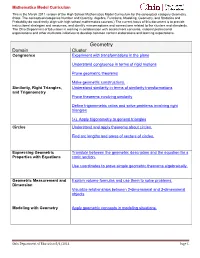
High School Geometry Model Curriculum Math.Pdf
Mathematics Model Curriculum This is the March 2011 version of the High School Mathematics Model Curriculum for the conceptual category Geometry. (Note: The conceptual categories Number and Quantity, Algebra, Functions, Modeling, Geometry, and Statistics and Probability do not directly align with high school mathematics courses.) The current focus of this document is to provide instructional strategies and resources, and identify misconceptions and connections related to the clusters and standards. The Ohio Department of Education is working in collaboration with assessment consortia, national professional organizations and other multistate initiatives to develop common content elaborations and learning expectations. Geometry Domain Cluster Congruence Experiment with transformations in the plane Understand congruence in terms of rigid motions Prove geometric theorems Make geometric constructions. Similarity, Right Triangles, Understand similarity in terms of similarity transformations and Trigonometry Prove theorems involving similarity Define trigonometric ratios and solve problems involving right triangles (+) Apply trigonometry to general triangles Circles Understand and apply theorems about circles. Find arc lengths and areas of sectors of circles. Expressing Geometric Translate between the geometric description and the equation for a Properties with Equations conic section. Use coordinates to prove simple geometric theorems algebraically. Geometric Measurement and Explain volume formulas and use them to solve problems. Dimension -

The Development of Thales Theorem Throughout History Slim Mrabet
The development of Thales theorem throughout history Slim Mrabet To cite this version: Slim Mrabet. The development of Thales theorem throughout history. Eleventh Congress of the European Society for Research in Mathematics Education, Utrecht University, Feb 2019, Utrecht, Netherlands. hal-02421913 HAL Id: hal-02421913 https://hal.archives-ouvertes.fr/hal-02421913 Submitted on 20 Dec 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. The development of Thales theorem throughout history Slim MRABET Carthage University, Tunisia; [email protected] Keywords: Thales theorem, similar triangles, transformation. Thales theorem may have different functionalities when using distances, algebraic measurements, or vectors. In addition to that, the utilization of a figure formed of secant lines and parallels or a figure relating to two similar triangles. The aim of this work is to categorize different formulations of Thales theorem and explain why in teaching we must know the appropriate mathematical environment related to each Thales Theorem statement. The analysis of many geometry books in history makes it possible to distinguish two points of view according to different forms, demonstrations and applications of this concept. The Euclidean point of view The general statement of Thales theorem shows us the idea to move from one triangle to another, moreover, the link with similar triangles (immediately following it and generally with similar figures) is a characteristic of this point of view. -
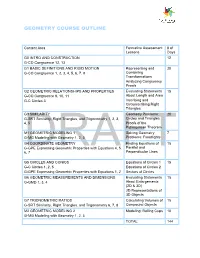
Geometry Course Outline
GEOMETRY COURSE OUTLINE Content Area Formative Assessment # of Lessons Days G0 INTRO AND CONSTRUCTION 12 G-CO Congruence 12, 13 G1 BASIC DEFINITIONS AND RIGID MOTION Representing and 20 G-CO Congruence 1, 2, 3, 4, 5, 6, 7, 8 Combining Transformations Analyzing Congruency Proofs G2 GEOMETRIC RELATIONSHIPS AND PROPERTIES Evaluating Statements 15 G-CO Congruence 9, 10, 11 About Length and Area G-C Circles 3 Inscribing and Circumscribing Right Triangles G3 SIMILARITY Geometry Problems: 20 G-SRT Similarity, Right Triangles, and Trigonometry 1, 2, 3, Circles and Triangles 4, 5 Proofs of the Pythagorean Theorem M1 GEOMETRIC MODELING 1 Solving Geometry 7 G-MG Modeling with Geometry 1, 2, 3 Problems: Floodlights G4 COORDINATE GEOMETRY Finding Equations of 15 G-GPE Expressing Geometric Properties with Equations 4, 5, Parallel and 6, 7 Perpendicular Lines G5 CIRCLES AND CONICS Equations of Circles 1 15 G-C Circles 1, 2, 5 Equations of Circles 2 G-GPE Expressing Geometric Properties with Equations 1, 2 Sectors of Circles G6 GEOMETRIC MEASUREMENTS AND DIMENSIONS Evaluating Statements 15 G-GMD 1, 3, 4 About Enlargements (2D & 3D) 2D Representations of 3D Objects G7 TRIONOMETRIC RATIOS Calculating Volumes of 15 G-SRT Similarity, Right Triangles, and Trigonometry 6, 7, 8 Compound Objects M2 GEOMETRIC MODELING 2 Modeling: Rolling Cups 10 G-MG Modeling with Geometry 1, 2, 3 TOTAL: 144 HIGH SCHOOL OVERVIEW Algebra 1 Geometry Algebra 2 A0 Introduction G0 Introduction and A0 Introduction Construction A1 Modeling With Functions G1 Basic Definitions and Rigid -

Similar Quadrilaterals Cui, Kadaveru, Lee, Maheshwari Page 1
Similar Quadrilaterals Cui, Kadaveru, Lee, Maheshwari Page 1 Similar Quadrilaterals Authors Guangqi Cui, Akshaj Kadaveru, Joshua Lee, Sagar Maheshwari Special thanks to Cosmin Pohoata and the AMSP Cornell 2014 Geometric Proofs Class B0 C0 B A A0 D0 C D Additional thanks to Justin Stevens and David Altizio for the LATEX Template Similar Quadrilaterals Cui, Kadaveru, Lee, Maheshwari Page 2 Contents 1 Introduction 3 2 Interesting Property 4 3 Example Problems 5 4 Practice Problems 11 Similar Quadrilaterals Cui, Kadaveru, Lee, Maheshwari Page 3 1 Introduction Similar quadrilaterals are a very useful but relatively unknown tool used to solve olympiad geometry problems. It usually goes unnoticed due to the confinement of geometric education to the geometry of the triangle and other conventional methods of problem solving. Also, it is only in very special cases where pairs of similar quadrilaterals exist, and proofs using these qualities usually shorten what would have otherwise been an unnecessarily long proof. The most common method of finding such quadrilaterals involves finding one pair of adjacent sides with identical ratios, and three pairs of congruent angles. We will call this SSAAA Similarity. 0 0 0 0 Example 1.1. (SSAAA Similarity) Two quadrilaterals ABCD and A B C D satisfy \A = AB BC A0, B = B0, C = C0, and = . Show that ABCD and A0B0C0D0 are similar. \ \ \ \ \ A0B0 B0C0 B0 C0 B A A0 D0 C D 0 0 0 0 0 0 Solution. Notice 4ABC and 4A B C are similar from SAS similarity. Therefore \C A D = 0 0 0 0 0 0 0 0 0 0 \A − \B A C = \A − \BAC = \CAD. -

∆ Congruence & Similarity
www.MathEducationPage.org Henri Picciotto Triangle Congruence and Similarity A Common-Core-Compatible Approach Henri Picciotto The Common Core State Standards for Mathematics (CCSSM) include a fundamental change in the geometry program in grades 8 to 10: geometric transformations, not congruence and similarity postulates, are to constitute the logical foundation of geometry at this level. This paper proposes an approach to triangle congruence and similarity that is compatible with this new vision. Transformational Geometry and the Common Core From the CCSSM1: The concepts of congruence, similarity, and symmetry can be understood from the perspective of geometric transformation. Fundamental are the rigid motions: translations, rotations, reflections, and combinations of these, all of which are here assumed to preserve distance and angles (and therefore shapes generally). Reflections and rotations each explain a particular type of symmetry, and the symmetries of an object offer insight into its attributes—as when the reflective symmetry of an isosceles triangle assures that its base angles are congruent. In the approach taken here, two geometric figures are defined to be congruent if there is a sequence of rigid motions that carries one onto the other. This is the principle of superposition. For triangles, congruence means the equality of all corresponding pairs of sides and all corresponding pairs of angles. During the middle grades, through experiences drawing triangles from given conditions, students notice ways to specify enough measures in a triangle to ensure that all triangles drawn with those measures are congruent. Once these triangle congruence criteria (ASA, SAS, and SSS) are established using rigid motions, they can be used to prove theorems about triangles, quadrilaterals, and other geometric figures. -

Document Similarity in Information Retrieval
Document Similarity in Information Retrieval Mausam (Based on slides of W. Arms, Thomas Hofmann, Ata Kaban, Melanie Martin) Standard Web Search Engine Architecture store documents, check for duplicates, extract links crawl the web DocIds create an user inverted query index Search inverted show results engine index To user servers Slide adapted from Marti Hearst / UC Berkeley] Indexing Subsystem documents Documents assign document IDs text break into tokens document numbers tokens stop list* and *field numbers non-stoplist stemming* tokens *Indicates optional stemmed term weighting* operation. terms terms with Index weights database Search Subsystem query parse query query tokens ranked document set stop list* non-stoplist tokens ranking* stemming* stemmed terms Boolean *Indicates retrieved operations* optional document set Index operation. relevant database document set Terms vs tokens • Terms are what results after tokenization and linguistic processing. – Examples • knowledge -> knowledg • The -> the • Removal of stop words Matching/Ranking of Textual Documents Major Categories of Methods 1. Exact matching (Boolean) 2. Ranking by similarity to query (vector space model) 3. Ranking of matches by importance of documents (PageRank) 4. Combination methods What happens in major search engines (Googlerank) Vector representation of documents and queries Why do this? • Represents a large space for documents • Compare – Documents – Documents with queries • Retrieve and rank documents with regards to a specific query - Enables methods of similarity All search engines do this. Boolean queries • Document is relevant to a query of the query itself is in the document. – Query blue and red brings back all documents with blue and red in them • Document is either relevant or not relevant to the query. -

SIMILARITY Euclidean Geometry Can Be
SIMILARITY OLEG VIRO Euclidean Geometry can be described as a study of the properties of geometric figures, but not all conceivable properties. Only the proper- ties which do not change under isometries deserve to be called geometric properties and studied in Euclidian Geometry. Some geometric properties are invariant under transformations that belong to wider classes. One such class of transformations is similarity transformations. Roughly they can be described as transformations preserving shapes, but changing scales: magnifying or contracting. The part of Euclidean Geometry that studies the geometric proper- ties unchanged by similarity transformations is called the similarity ge- ometry. Similarity geometry can be introduced in a number of different ways. The most straightforward of them is based on the notion of ratio of segments. The similarity geometry is an integral part of Euclidean Geometry. However, its main notions emerge in traditional presentations of Eu- clidean Geometry (in particular in the textbook by Hadamard that we use) in a very indirect way. Below it is shown how this can be done ac- cording to the standards of modern mathematics. But first, in Sections 1 - 4, the traditional definitions for ratio of segments and the Euclidean distance are summarized. 1. Ratio of commensurable segments. If a segment CD can be obtained by summing up of n copies of a segment AB, then we say CD AB 1 that = n and = . AB CD n If for segments AB and CD there exists a segment EF and natural AB CD numbers p and q such that = p and = q, then AB and CD are EF EF AB p said to be commensurable, is defined as and the segment EF is CD q called a common measure of AB and CD. -

Chapter 4 Euclidean Geometry
Chapter 4 Euclidean Geometry Based on previous 15 axioms, The parallel postulate for Euclidean geometry is added in this chapter. 4.1 Euclidean Parallelism, Existence of Rectangles De¯nition 4.1 Two distinct lines ` and m are said to be parallel ( and we write `km) i® they lie in the same plane and do not meet. Terminologies: 1. Transversal: a line intersecting two other lines. 2. Alternate interior angles 3. Corresponding angles 4. Interior angles on the same side of transversal 56 Yi Wang Chapter 4. Euclidean Geometry 57 Theorem 4.2 (Parallelism in absolute geometry) If two lines in the same plane are cut by a transversal to that a pair of alternate interior angles are congruent, the lines are parallel. Remark: Although this theorem involves parallel lines, it does not use the parallel postulate and is valid in absolute geometry. Proof: Assume to the contrary that the two lines meet, then use Exterior Angle Inequality to draw a contradiction. 2 The converse of above theorem is the Euclidean Parallel Postulate. Euclid's Fifth Postulate of Parallels If two lines in the same plane are cut by a transversal so that the sum of the measures of a pair of interior angles on the same side of the transversal is less than 180, the lines will meet on that side of the transversal. In e®ect, this says If m\1 + m\2 6= 180; then ` is not parallel to m Yi Wang Chapter 4. Euclidean Geometry 58 It's contrapositive is If `km; then m\1 + m\2 = 180( or m\2 = m\3): Three possible notions of parallelism Consider in a single ¯xed plane a line ` and a point P not on it. -

Comparison Dimensions and Similarity: Addressing Individual Heterogeneity
DISCUSSION PAPER SERIES IZA DP No. 12355 Comparison Dimensions and Similarity: Addressing Individual Heterogeneity Pavel Jelnov MAY 2019 DISCUSSION PAPER SERIES IZA DP No. 12355 Comparison Dimensions and Similarity: Addressing Individual Heterogeneity Pavel Jelnov Leibniz University Hannover and IZA MAY 2019 Any opinions expressed in this paper are those of the author(s) and not those of IZA. Research published in this series may include views on policy, but IZA takes no institutional policy positions. The IZA research network is committed to the IZA Guiding Principles of Research Integrity. The IZA Institute of Labor Economics is an independent economic research institute that conducts research in labor economics and offers evidence-based policy advice on labor market issues. Supported by the Deutsche Post Foundation, IZA runs the world’s largest network of economists, whose research aims to provide answers to the global labor market challenges of our time. Our key objective is to build bridges between academic research, policymakers and society. IZA Discussion Papers often represent preliminary work and are circulated to encourage discussion. Citation of such a paper should account for its provisional character. A revised version may be available directly from the author. ISSN: 2365-9793 IZA – Institute of Labor Economics Schaumburg-Lippe-Straße 5–9 Phone: +49-228-3894-0 53113 Bonn, Germany Email: [email protected] www.iza.org IZA DP No. 12355 MAY 2019 ABSTRACT Comparison Dimensions and Similarity: Addressing Individual Heterogeneity How many comparison dimensions individuals consider when they are asked to judge how similar two different objects are? I address individual heterogeneity in the number of comparison dimensions with data from a laboratory experiment. -

Similarity Measures Similarity and Dissimilarity Are Important Because
Similarity Measures Similarity and dissimilarity are important because they are used by a number of data mining techniques, such as clustering nearest neighbor classification and anomaly detection. The term proximity is used to refer to either similarity or dissimilarity. Definitions: The similarity between two objects is a numeral measure of the degree to which the two objects are alike. Consequently, similarities are higher for pairs of objects that are more alike. Similarities are usually non-negative and are often between 0 (no similarity) and 1(complete similarity). The dissimilarity between two objects is the numerical measure of the degree to which the two objects are different. Dissimilarity is lower for more similar pairs of objects. Frequently, the term distance is used as a synonym for dissimilarity. Dissimilarities sometimes fall in the interval [0,1], but it is also common for them to range from 0 to Proximity Measures: Proximity measures, especially similarities, are defined to have values in the interval [0,1]. If the similarity between objects can range from 1 (not at all similar) to 10 (completely similar), we can make them fall into the range [0,1] by using the formula: s’=(s-1)/9, where s and s’ are the original and the new similarity values, respectively. The more general case, s’ is calculated as s’=(s-min_s)/(max_s-min_s), where min_s and max_s are the minimum and maximum similarity values respectively. Likewise, dissimilarity measures with a finite range can be mapped to the interval [0,1] by using the formula d’=(d-min_d)/(max_d- min_d). If the proximity measure originally takes values in the interval [0, ], then we usually use the formula: d’= d/(1+d) for such cases and bring the dissimilarity measure between [0,1]. -

SIMILARITY Euclidean Geometry Can Be Described As a Study of The
SIMILARITY BASED ON NOTES BY OLEG VIRO, REVISED BY OLGA PLAMENEVSKAYA Euclidean Geometry can be described as a study of the properties of geometric figures, but not all kinds of conceivable properties. Only the properties which do not change under isometries deserve to be called geometric properties and studied in Euclidian Geometry. Some geometric properties are invariant under transformations that belongto wider classes. One such class of transformations is similarity transformations. Roughly they can be described as transformations preserving shapes, but changing scales: magnifying or contracting. The part of Euclidean Geometry that studies the geometric properties unchanged by similarity transformations is called the similarity geometry. Similarity geometry can be introduced in a number of different ways. The most straightforward of them is based on the notion of ratio of segments. The similarity geometry is an integral part of Euclidean Geometry. In fact, there is no interesting phenomenon that belong to Euclidean Geometry, but does not survive a rescaling. In this sense, the whole Euclidean Geometry can be considered through the glass of the similarity geometry. Moreover, all the results of Euclidean Geometry concerning relations among distances are obtained using similarity transformations. However, main notions of the similarity geometry emerge in traditional presenta- tions of Euclidean Geometry (in particular, in the Kiselev textbook) in a very indirect way. Below it is shown how this can be done more naturally, according to the stan- dards of modern mathematics. But first, in Sections 1 - 4, the traditional definitions for ratio of segments and the Euclidean distance are summarized. 1. Ratio of commensurable segments.