A Review of Gaussian Random Matrices
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Chapter 2 a Short Review of Matrix Algebra
Chapter 2 A short review of matrix algebra 2.1 Vectors and vector spaces Definition 2.1.1. A vector a of dimension n is a collection of n elements typically written as ⎛ ⎞ ⎜ a1 ⎟ ⎜ ⎟ ⎜ ⎟ ⎜ a2 ⎟ a = ⎜ ⎟ =(ai)n. ⎜ . ⎟ ⎝ . ⎠ an Vectors of length 2 (two-dimensional vectors) can be thought of points in 33 BIOS 2083 Linear Models Abdus S. Wahed the plane (See figures). Chapter 2 34 BIOS 2083 Linear Models Abdus S. Wahed Figure 2.1: Vectors in two and three dimensional spaces (-1.5,2) (1, 1) (1, -2) x1 (2.5, 1.5, 0.95) x2 (0, 1.5, 0.95) x3 Chapter 2 35 BIOS 2083 Linear Models Abdus S. Wahed • A vector with all elements equal to zero is known as a zero vector and is denoted by 0. • A vector whose elements are stacked vertically is known as column vector whereas a vector whose elements are stacked horizontally will be referred to as row vector. (Unless otherwise mentioned, all vectors will be referred to as column vectors). • A row vector representation of a column vector is known as its trans- T pose. We will use⎛ the⎞ notation ‘ ’or‘ ’ to indicate a transpose. For ⎜ a1 ⎟ ⎜ ⎟ ⎜ a2 ⎟ ⎜ ⎟ T instance, if a = ⎜ ⎟ and b =(a1 a2 ... an), then we write b = a ⎜ . ⎟ ⎝ . ⎠ an or a = bT . • Vectors of same dimension are conformable to algebraic operations such as additions and subtractions. Sum of two or more vectors of dimension n results in another n-dimensional vector with elements as the sum of the corresponding elements of summand vectors. -

Facts from Linear Algebra
Appendix A Facts from Linear Algebra Abstract We introduce the notation of vector and matrices (cf. Section A.1), and recall the solvability of linear systems (cf. Section A.2). Section A.3 introduces the spectrum σ(A), matrix polynomials P (A) and their spectra, the spectral radius ρ(A), and its properties. Block structures are introduced in Section A.4. Subjects of Section A.5 are orthogonal and orthonormal vectors, orthogonalisation, the QR method, and orthogonal projections. Section A.6 is devoted to the Schur normal form (§A.6.1) and the Jordan normal form (§A.6.2). Diagonalisability is discussed in §A.6.3. Finally, in §A.6.4, the singular value decomposition is explained. A.1 Notation for Vectors and Matrices We recall that the field K denotes either R or C. Given a finite index set I, the linear I space of all vectors x =(xi)i∈I with xi ∈ K is denoted by K . The corresponding square matrices form the space KI×I . KI×J with another index set J describes rectangular matrices mapping KJ into KI . The linear subspace of a vector space V spanned by the vectors {xα ∈V : α ∈ I} is denoted and defined by α α span{x : α ∈ I} := aαx : aα ∈ K . α∈I I×I T Let A =(aαβ)α,β∈I ∈ K . Then A =(aβα)α,β∈I denotes the transposed H matrix, while A =(aβα)α,β∈I is the adjoint (or Hermitian transposed) matrix. T H Note that A = A holds if K = R . Since (x1,x2,...) indicates a row vector, T (x1,x2,...) is used for a column vector. -

On the Distance Spectra of Graphs
On the distance spectra of graphs Ghodratollah Aalipour∗ Aida Abiad† Zhanar Berikkyzy‡ Jay Cummings§ Jessica De Silva¶ Wei Gaok Kristin Heysse‡ Leslie Hogben‡∗∗ Franklin H.J. Kenter†† Jephian C.-H. Lin‡ Michael Tait§ October 5, 2015 Abstract The distance matrix of a graph G is the matrix containing the pairwise distances between vertices. The distance eigenvalues of G are the eigenvalues of its distance matrix and they form the distance spectrum of G. We determine the distance spectra of halved cubes, double odd graphs, and Doob graphs, completing the determination of distance spectra of distance regular graphs having exactly one positive distance eigenvalue. We characterize strongly regular graphs having more positive than negative distance eigenvalues. We give examples of graphs with few distinct distance eigenvalues but lacking regularity properties. We also determine the determinant and inertia of the distance matrices of lollipop and barbell graphs. Keywords. distance matrix, eigenvalue, distance regular graph, Kneser graph, double odd graph, halved cube, Doob graph, lollipop graph, barbell graph, distance spectrum, strongly regular graph, optimistic graph, determinant, inertia, graph AMS subject classifications. 05C12, 05C31, 05C50, 15A15, 15A18, 15B48, 15B57 1 Introduction The distance matrix (G) = [d ] of a graph G is the matrix indexed by the vertices of G where d = d(v , v ) D ij ij i j is the distance between the vertices vi and vj , i.e., the length of a shortest path between vi and vj . Distance matrices were introduced in the study of a data communication problem in [16]. This problem involves finding appropriate addresses so that a message can move efficiently through a series of loops from its origin to its destination, choosing the best route at each switching point. -

Spectra of Graphs
Spectra of graphs Andries E. Brouwer Willem H. Haemers 2 Contents 1 Graph spectrum 11 1.1 Matricesassociatedtoagraph . 11 1.2 Thespectrumofagraph ...................... 12 1.2.1 Characteristicpolynomial . 13 1.3 Thespectrumofanundirectedgraph . 13 1.3.1 Regulargraphs ........................ 13 1.3.2 Complements ......................... 14 1.3.3 Walks ............................. 14 1.3.4 Diameter ........................... 14 1.3.5 Spanningtrees ........................ 15 1.3.6 Bipartitegraphs ....................... 16 1.3.7 Connectedness ........................ 16 1.4 Spectrumofsomegraphs . 17 1.4.1 Thecompletegraph . 17 1.4.2 Thecompletebipartitegraph . 17 1.4.3 Thecycle ........................... 18 1.4.4 Thepath ........................... 18 1.4.5 Linegraphs .......................... 18 1.4.6 Cartesianproducts . 19 1.4.7 Kronecker products and bipartite double. 19 1.4.8 Strongproducts ....................... 19 1.4.9 Cayleygraphs......................... 20 1.5 Decompositions............................ 20 1.5.1 Decomposing K10 intoPetersengraphs . 20 1.5.2 Decomposing Kn into complete bipartite graphs . 20 1.6 Automorphisms ........................... 21 1.7 Algebraicconnectivity . 22 1.8 Cospectralgraphs .......................... 22 1.8.1 The4-cube .......................... 23 1.8.2 Seidelswitching. 23 1.8.3 Godsil-McKayswitching. 24 1.8.4 Reconstruction ........................ 24 1.9 Verysmallgraphs .......................... 24 1.10 Exercises ............................... 25 3 4 CONTENTS 2 Linear algebra 29 2.1 -

EXPLICIT BOUNDS for the PSEUDOSPECTRA of VARIOUS CLASSES of MATRICES and OPERATORS Contents 1. Introduction 2 2. Pseudospectra 2
EXPLICIT BOUNDS FOR THE PSEUDOSPECTRA OF VARIOUS CLASSES OF MATRICES AND OPERATORS FEIXUE GONG1, OLIVIA MEYERSON2, JEREMY MEZA3, ABIGAIL WARD4 MIHAI STOICIU5 (ADVISOR) SMALL 2014 - MATHEMATICAL PHYSICS GROUP N×N Abstract. We study the "-pseudospectra σ"(A) of square matrices A ∈ C . We give a complete characterization of the "-pseudospectrum of any 2×2 matrix and describe the asymptotic behavior (as " → 0) of σ"(A) for any square matrix A. We also present explicit upper and lower bounds for the "-pseudospectra of bidiagonal and tridiagonal matrices, as well as for finite rank operators. Contents 1. Introduction 2 2. Pseudospectra 2 2.1. Motivation and Definitions 2 2.2. Normal Matrices 6 2.3. Non-normal Diagonalizable Matrices 7 2.4. Non-diagonalizable Matrices 8 3. Pseudospectra of 2 2 Matrices 10 3.1. Non-diagonalizable× 2 2 Matrices 10 3.2. Diagonalizable 2 2 Matrices× 12 4. Asymptotic Union of Disks× Theorem 15 5. Pseudospectra of N N Jordan Block 20 6. Pseudospectra of bidiagonal× matrices 22 6.1. Asymptotic Bounds for Periodic Bidiagonal Matrices 22 6.2. Examples of Bidiagonal Matrices 29 7. Tridiagonal Matrices 32 Date: October 30, 2014. 1;2;5Williams College 3Carnegie Mellon University 4University of Chicago. 1 2 GONG, MEYERSON, MEZA, WARD, STOICIU 8. Finite Rank operators 33 References 35 1. Introduction The pseudospectra of matrices and operators is an important mathematical object that has found applications in various areas of mathematics: linear algebra, func- tional analysis, numerical analysis, and differential equations. An overview of the main results on pseudospectra can be found in [14]. -
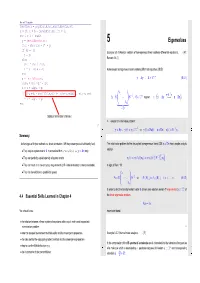
Eigenvalues Rho1 = Rho; Rho = R' * Y; If (I == 1) Example 5.0.1 (Analytic Solution of Homogeneous Linear Ordinary Differential Equations)
MATLAB PCG algorithm function x = pcg(Afun,b,tol,maxit,Binvfun,x0) x = x0; r = b - feval(Afun,x); rho = 1; for i = 1 : maxit y = feval(Binvfun,r); 5 Eigenvalues rho1 = rho; rho = r' * y; if (i == 1) Example 5.0.1 (Analytic solution of homogeneous linear ordinary differential equations). → [47, p = y; else Remark 5.6.1] beta = rho / rho1; p = y + beta * p; Autonomous homogeneous linear ordinary differential equation (ODE): end q = feval(Afun,p); y˙ = Ay , A ∈ Cn,n . (5.0.1) alpha = rho /(p' * q); x = x + alpha * p; λ 1 if (norm(b - evalf(Afun,x)) <= tolb*norm(b)) , return; end 1 z=S− y A = S ... S−1 , S ∈ Cn,n regular =⇒ y˙ = Ay ←→ z˙ = Dz . r = r - alpha * q; end λn =:D Dubious termination criterion ! | {z } ➣ solution of initial value problem: △ n −1 4º¿ y˙ = Ay , y(0) = y0 ∈ C ⇒ y(t) = Sz(t) , z˙ = Dz , z(0) = S y0 . 5º¼ Summary: Ôº ¾77 Ôº ¾79 Advantages of Krylov methods vs. direct elimination (, IF they converge at all/sufficiently fast). The initial value problem for the decoupled homogeneous linear ODE z˙ = Dz has a simple analytic • They require system matrix A in procedural form y=evalA(x) ↔ y = Ax only. solution −1 T • They can perfectly exploit sparsity of system matrix. zi(t) = exp(λit)(z0)i = exp(λit) (S )i,:y0 . • They can cash in on low accuracy requirements (, IF viable termination criterion available). In light of Rem. ??: • They can benefit from a good initial guess. λ1 A = S ... S−1 ⇔ A (S) = λ (S) i = 1,..., n . -

Symmetric Rank-One Updates from Partial Spectrum with an Application to Out-Of-Sample Extension
Symmetric rank-one updates from partial spectrum with an application to out-of-sample extension Roy Mitz∗ 1, Nir Sharon† 1, and Yoel Shkolnisky‡1 1School of Mathematical Sciences, Tel-Aviv University, Tel-Aviv, Israel Abstract Rank-one update of the spectrum of a matrix is a fundamental problem in classical perturbation theory. In this paper, we consider its variant where only part of the spectrum is known. We address this variant using an efficient scheme for updating the known eigenpairs with guaranteed error bounds. Then, we apply our scheme to the extension of the top eigenvectors of the graph Laplacian to a new data sample. In particular, we model this extension as a perturbation problem and show how to solve it using our rank-one updating scheme. We provide a theoretical analysis of this extension method, and back it up with numerical results that illustrate its advantages. 1 Introduction The last few decades have witnessed the emergence of various algorithms that require the calculation of the eigendecomposition of matrices. A few known examples are Google's PageRank [18], PCA [21], Laplacian eigenmaps [1], LLE [20] and MDS [5]. Since datasets nowadays may contain tens of millions of data points, an efficient calculation of the eigendecomposition becomes fundamental. In many scenarios, only part of the eigendecomposition, i.e., only the leading eigenvalues and eigenvectors, can or need to be calculated. While algorithms for eigendecomposition, such as the Lanczos algorithm and some variants of SVD, are designed especially for this task, they still require a hefty amount of calculations. A natural question that arises in such cases is how to update the eigendecomposition of a matrix given its partial arXiv:1710.02774v4 [math.NA] 8 Jul 2019 eigendecomposition and some \small" perturbation to it, without repeating the entire decomposition again. -

October 30, 2013 SPECTRAL PROPERTIES of SELF-ADJOINT MATRICES
October 30, 2013 SPECTRAL PROPERTIES OF SELF-ADJOINT MATRICES RODICA D. COSTIN Contents 1. Review 3 1.1. The spectrum of a matrix 3 1.2. Brief overview of previous results 3 1.3. More on similar matrices 4 2. Self-adjoint matrices 4 2.1. Definitions 4 2.2. Self-adjoint matrices are diagonalizable I 5 2.3. Further properties of unitary matrices 7 2.4. An alternative look at the proof of Theorem 16 8 2.5. Triangularization by conjugation using a unitary matrix 8 2.6. All self-adjoint matrices are diagonalizable II 10 2.7. Normal matrices 10 2.8. Generic matrices (or: ”beware of roundo↵errors”) 11 2.9. Anti-self-adjoint (skew-symmetric, skew-Hermitian) matrices 13 2.10. Application to linear di↵erential equations 13 2.11. Diagonalization of unitary matrices 14 3. Quadratic forms and Positive definite matrices 15 3.1. Quadratic forms 15 3.2. Critical points of functions of several variables. 18 3.3. Positive definite matrices 19 3.4. Negative definite, semidefinite and indefinite matrices 20 3.5. Applications to critical points of several variables 21 3.6. Application to di↵erential equations: Lyapunov functions 22 3.7. Solving linear systems by minimization 25 3.8. Generalized eigenvalue problems 26 4. The Rayleigh’s principle and the minimax principle for the eigenvalues of a self-adjoint matrix 28 4.1. The Rayleigh’s quotient 28 4.2. Extrema of the Rayleigh’s quotient 28 4.3. The minimax principle 30 4.4. The minimax principle for the generalized eigenvalue problem. -

Eigenvalue Estimates Via Pseudospectra †
mathematics Article Eigenvalue Estimates via Pseudospectra † Georgios Katsouleas *,‡, Vasiliki Panagakou ‡ and Panayiotis Psarrakos ‡ Department of Mathematics, Zografou Campus, National Technical University of Athens, 15773 Athens, Greece; [email protected] (V.P.); [email protected] (P.P.) * Correspondence: [email protected] † This paper is dedicated to Mr. Constantin M. Petridi. ‡ These authors contributed equally to this work. × Abstract: In this note, given a matrix A 2 Cn n (or a general matrix polynomial P(z), z 2 C) and 2 f g an arbitrary scalar l0 C, we show how to define a sequence mk k2N which converges to some element of its spectrum. The scalar l0 serves as initial term (m0 = l0), while additional terms are constructed through a recursive procedure, exploiting the fact that each term mk of this sequence is in fact a point lying on the boundary curve of some pseudospectral set of A (or P(z)). Then, the next term in the sequence is detected in the direction which is normal to this curve at the point mk. Repeating the construction for additional initial points, it is possible to approximate peripheral eigenvalues, localize the spectrum and even obtain spectral enclosures. Hence, as a by-product of our method, a computationally cheap procedure for approximate pseudospectra computations emerges. An advantage of the proposed approach is that it does not make any assumptions on the location of the spectrum. The fact that all computations are performed on some dynamically chosen locations on the complex plane which converge to the eigenvalues, rather than on a large number of predefined points on a rigid grid, can be used to accelerate conventional grid algorithms. -

Types of Convergence of Matrices by Olga Pryporova a Dissertation
Types of convergence of matrices by Olga Pryporova A dissertation submitted to the graduate faculty in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Major: Mathematics Program of Study Committee: Leslie Hogben, Major Professor Maria Axenovich Wolfgang Kliemann Yiu Poon Paul Sacks Iowa State University Ames, Iowa 2009 ii TABLE OF CONTENTS CHAPTER 1. GENERAL INTRODUCTION . 1 1.1 Introduction . 1 1.2 Thesis Organization . 3 1.3 Literature Review . 4 CHAPTER 2. COMPLEX D CONVERGENCE AND DIAGONAL CONVERGENCE OF MATRICES . 10 2.1 Introduction . 10 2.2 When Boundary Convergence Implies Diagonal Convergence . 14 2.3 Examples of 2 × 2 and 3 × 3 Matrices . 24 CHAPTER 3. QUALITATIVE CONVERGENCE OF MATRICES . 27 3.1 Introduction . 27 3.2 Types of Potential Convergence . 30 3.3 Characterization of Potential Absolute and Potential Diagonal Convergence . 34 3.4 Results on Potential Convergence and Potential D Convergence . 35 3.4.1 2 × 2 Modulus Patterns . 38 3.4.2 Some Examples of 3 × 3 Modulus Patterns . 41 CHAPTER 4. GENERAL CONCLUSIONS . 44 4.1 General Discussion . 44 4.2 Recommendations for Future Research . 46 BIBLIOGRAPHY . 47 ACKNOWLEDGEMENTS . 51 1 CHAPTER 1. GENERAL INTRODUCTION In this work properties of the spectra of matrices that play an important role in applications to stability of dynamical systems are studied. 1.1 Introduction A (square) complex matrix is stable (also referred to as Hurwitz stable or continuous time stable) if all its eigenvalues lie in the open left half plane of the complex plane. It is well known (see, for example, [29, p. -
The Normalized Distance Laplacian Received July 1, 2020; Accepted October 17, 2020
Spec. Matrices 2021; 9:1–18 Research Article Open Access Special Issue: Combinatorial Matrices Carolyn Reinhart* The normalized distance Laplacian https://doi.org/10.1515/spma-2020-0114 Received July 1, 2020; accepted October 17, 2020 Abstract: The distance matrix D(G) of a connected graph G is the matrix containing the pairwise distances between vertices. The transmission of a vertex vi in G is the sum of the distances from vi to all other vertices and T(G) is the diagonal matrix of transmissions of the vertices of the graph. The normalized distance Lapla- cian, DL(G) = I−T(G)−1/2D(G)T(G)−1/2, is introduced. This is analogous to the normalized Laplacian matrix, L(G) = I − D(G)−1/2A(G)D(G)−1/2, where D(G) is the diagonal matrix of degrees of the vertices of the graph and A(G) is the adjacency matrix. Bounds on the spectral radius of DL and connections with the normalized Laplacian matrix are presented. Twin vertices are used to determine eigenvalues of the normalized distance Laplacian. The distance generalized characteristic polynomial is dened and its properties established. Fi- nally, DL-cospectrality and lack thereof are determined for all graphs on 10 and fewer vertices, providing evidence that the normalized distance Laplacian has fewer cospectral pairs than other matrices. Keywords: Normalized Laplacian, Distance Matrices, Cospectrality, Generalized characteristic polynomial MSC: 05C50, 05C12, 15A18 1 Introduction Spectral graph theory is the study of matrices dened in terms of a graph, specically relating the eigenval- ues of the matrix to properties of the graph. -

Spectral Properties of Self-Adjoint Matrices
November 15, 2013 SPECTRAL PROPERTIES OF SELF-ADJOINT MATRICES RODICA D. COSTIN Contents 1. Review 3 1.1. The spectrum of a matrix 3 1.2. Brief overview of previous results 3 1.3. More on similar matrices 4 2. Self-adjoint matrices 4 2.1. Definitions 4 2.2. Self-adjoint matrices are diagonalizable I 5 2.3. Further properties of unitary matrices 7 2.4. An alternative look at the proof of Theorem 16 8 2.5. Triangularization by conjugation using a unitary matrix 8 2.6. All self-adjoint matrices are diagonalizable II 10 2.7. Normal matrices 10 2.8. Generic matrices (or: "beware of roundoff errors") 11 2.9. Anti-self-adjoint (skew-symmetric, skew-Hermitian) matrices 13 2.10. Application to linear differential equations 13 2.11. Diagonalization of unitary matrices 14 3. Quadratic forms and Positive definite matrices 15 3.1. Quadratic forms 15 3.2. Critical points of functions of several variables. 18 3.3. Positive definite matrices 20 3.4. Negative definite, semidefinite and indefinite matrices 21 3.5. Applications to critical points of several variables 22 3.6. Application to differential equations: Lyapunov functions 23 3.7. Solving linear systems by minimization 25 3.8. Generalized eigenvalue problems 26 4. The Rayleigh's principle. The minimax theorem for the eigenvalues of a self-adjoint matrix 28 4.1. The Rayleigh's quotient 28 4.2. Extrema of the Rayleigh's quotient 28 4.3. The minimax principle 30 4.4. The minimax principle for the generalized eigenvalue problem. 33 5.