Ruud Scharn Validating a New Analytical Framework for Including
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Field Release of the Leaf-Feeding Moth, Hypena Opulenta (Christoph)
United States Department of Field release of the leaf-feeding Agriculture moth, Hypena opulenta Marketing and Regulatory (Christoph) (Lepidoptera: Programs Noctuidae), for classical Animal and Plant Health Inspection biological control of swallow- Service worts, Vincetoxicum nigrum (L.) Moench and V. rossicum (Kleopow) Barbarich (Gentianales: Apocynaceae), in the contiguous United States. Final Environmental Assessment, August 2017 Field release of the leaf-feeding moth, Hypena opulenta (Christoph) (Lepidoptera: Noctuidae), for classical biological control of swallow-worts, Vincetoxicum nigrum (L.) Moench and V. rossicum (Kleopow) Barbarich (Gentianales: Apocynaceae), in the contiguous United States. Final Environmental Assessment, August 2017 Agency Contact: Colin D. Stewart, Assistant Director Pests, Pathogens, and Biocontrol Permits Plant Protection and Quarantine Animal and Plant Health Inspection Service U.S. Department of Agriculture 4700 River Rd., Unit 133 Riverdale, MD 20737 Non-Discrimination Policy The U.S. Department of Agriculture (USDA) prohibits discrimination against its customers, employees, and applicants for employment on the bases of race, color, national origin, age, disability, sex, gender identity, religion, reprisal, and where applicable, political beliefs, marital status, familial or parental status, sexual orientation, or all or part of an individual's income is derived from any public assistance program, or protected genetic information in employment or in any program or activity conducted or funded by the Department. (Not all prohibited bases will apply to all programs and/or employment activities.) To File an Employment Complaint If you wish to file an employment complaint, you must contact your agency's EEO Counselor (PDF) within 45 days of the date of the alleged discriminatory act, event, or in the case of a personnel action. -

Growing Plants for Hawaiian Lei ‘A‘Ali‘I
6 Growing Plants for Hawaiian Lei ‘a‘ali‘i OTHER COMMON NAMES: ‘a‘ali‘i kū range of habitats from dunes at sea makani, ‘a‘ali‘i kū ma kua, kū- level up through leeward and dry makani, hop bush, hopseed bush forests and to the highest peaks SCIENTIFIC NAME: Dodonaea viscosa CURRENT STATUS IN THE WILD IN HAWAI‘I: common FAMILY: Sapindaceae (soapberry family) CULTIVARS: female cultivars such as ‘Purpurea’ and ‘Saratoga’ have NATURAL SETTING/LOCATION: indigenous, been selected for good fruit color pantropical species, found on all the main Hawaiian Islands except Kaho‘olawe; grows in a wide Growing your own PROPAGATION FORM: seeds; semi-hardwood cuttings or air layering for selected color forms PREPLANTING TREATMENT: step on seed capsule to release small, round, black seeds, or use heavy gloves and rub capsules vigorously between hands; put seeds in water that has been brought to a boil and removed from heat, soak for about 24 hours; if seeds start to swell, sow imme- diately; discard floating, nonviable seeds; use strong rooting hormone on cuttings TEMPERATURE: PLANTING DEPTH: sow seeds ¼" deep in tolerates dry heat; tem- after fruiting period to shape or keep medium; insert base of cutting 1–2" perature 32–90°F short; can be shaped into a small tree or maintained as a shrub, hedge, or into medium ELEVATION: 10–7700' espalier (on a trellis) GERMINATION TIME: 2–4 weeks SALT TOLERANCE: good (moderate at SPECIAL CULTURAL HINTS: male and female CUTTING ROOTING TIME: 1½–3 months higher elevations) plants are separate, although bisex- WIND RESISTANCE: -

Sistema De Clasificación Artificial De Las Magnoliatas Sinántropas De Cuba
Sistema de clasificación artificial de las magnoliatas sinántropas de Cuba. Pedro Pablo Herrera Oliver Tesis doctoral de la Univerisdad de Alicante. Tesi doctoral de la Universitat d'Alacant. 2007 Sistema de clasificación artificial de las magnoliatas sinántropas de Cuba. Pedro Pablo Herrera Oliver PROGRAMA DE DOCTORADO COOPERADO DESARROLLO SOSTENIBLE: MANEJOS FORESTAL Y TURÍSTICO UNIVERSIDAD DE ALICANTE, ESPAÑA UNIVERSIDAD DE PINAR DEL RÍO, CUBA TESIS EN OPCIÓN AL GRADO CIENTÍFICO DE DOCTOR EN CIENCIAS SISTEMA DE CLASIFICACIÓN ARTIFICIAL DE LAS MAGNOLIATAS SINÁNTROPAS DE CUBA Pedro- Pabfc He.r retira Qltver CUBA 2006 Tesis doctoral de la Univerisdad de Alicante. Tesi doctoral de la Universitat d'Alacant. 2007 Sistema de clasificación artificial de las magnoliatas sinántropas de Cuba. Pedro Pablo Herrera Oliver PROGRAMA DE DOCTORADO COOPERADO DESARROLLO SOSTENIBLE: MANEJOS FORESTAL Y TURÍSTICO UNIVERSIDAD DE ALICANTE, ESPAÑA Y UNIVERSIDAD DE PINAR DEL RÍO, CUBA TESIS EN OPCIÓN AL GRADO CIENTÍFICO DE DOCTOR EN CIENCIAS SISTEMA DE CLASIFICACIÓN ARTIFICIAL DE LAS MAGNOLIATAS SINÁNTROPAS DE CUBA ASPIRANTE: Lie. Pedro Pablo Herrera Oliver Investigador Auxiliar Centro Nacional de Biodiversidad Instituto de Ecología y Sistemática Ministerio de Ciencias, Tecnología y Medio Ambiente DIRECTORES: CUBA Dra. Nancy Esther Ricardo Ñapóles Investigador Titular Centro Nacional de Biodiversidad Instituto de Ecología y Sistemática Ministerio de Ciencias, Tecnología y Medio Ambiente ESPAÑA Dr. Andreu Bonet Jornet Piiofesjar Titular Departamento de EGdfegfe Universidad! dte Mearte CUBA 2006 Tesis doctoral de la Univerisdad de Alicante. Tesi doctoral de la Universitat d'Alacant. 2007 Sistema de clasificación artificial de las magnoliatas sinántropas de Cuba. Pedro Pablo Herrera Oliver I. INTRODUCCIÓN 1 II. ANTECEDENTES 6 2.1 Historia de los esquemas de clasificación de las especies sinántropas (1903-2005) 6 2.2 Historia del conocimiento de las plantas sinantrópicas en Cuba 14 III. -

Pu'u Wa'awa'a Biological Assessment
PU‘U WA‘AWA‘A BIOLOGICAL ASSESSMENT PU‘U WA‘AWA‘A, NORTH KONA, HAWAII Prepared by: Jon G. Giffin Forestry & Wildlife Manager August 2003 STATE OF HAWAII DEPARTMENT OF LAND AND NATURAL RESOURCES DIVISION OF FORESTRY AND WILDLIFE TABLE OF CONTENTS TITLE PAGE ................................................................................................................................. i TABLE OF CONTENTS ............................................................................................................. ii GENERAL SETTING...................................................................................................................1 Introduction..........................................................................................................................1 Land Use Practices...............................................................................................................1 Geology..................................................................................................................................3 Lava Flows............................................................................................................................5 Lava Tubes ...........................................................................................................................5 Cinder Cones ........................................................................................................................7 Soils .......................................................................................................................................9 -

Responses of Ornamental Mussaenda Species Stem Cuttings to Varying Concentrations of Naphthalene Acetic Acid Phytohormone Application
GSC Biological and Pharmaceutical Sciences, 2017, 01(01), 020–024 Available online at GSC Online Press Directory GSC Biological and Pharmaceutical Sciences e-ISSN: 2581-3250, CODEN (USA): GBPSC2 Journal homepage: https://www.gsconlinepress.com/journals/gscbps (RESEARCH ARTICLE) Responses of ornamental Mussaenda species stem cuttings to varying concentrations of naphthalene acetic acid phytohormone application Ogbu Justin U. 1*, Okocha Otah I. 2 and Oyeleye David A. 3 1 Department of Horticulture and Landscape technology, Federal College of Agriculture (FCA), Ishiagu 491105 Nigeria. 2 Department of Horticulture technology, AkanuIbiam Federal Polytechnic, Unwana Ebonyi state Nigeria. 3 Department of Agricultural technology, Federal College of Agriculture (FCA), Ishiagu 491105 Nigeria Publication history: Received on 28 August 2017; revised on 03 October 2017; accepted on 09 October 2017 https://doi.org/10.30574/gscbps.2017.1.1.0009 Abstract This study evaluated the rooting and sprouting responses of four ornamental Mussaendas species (Flag bush) stem cuttings to treatment with varying concentrations of 1-naphthalene acetic acid (NAA). Species evaluated include Mussaenda afzelii (wild), M. erythrophylla, M. philippica and Pseudomussaenda flava. Different concentrations of NAA phytohormone were applied to the cuttings grown in mixed river sand and saw dust (1:1; v/v); and laid out in a 4 x 4 factorial experiment in completely randomized design (CRD; r=4). Results showed that increasing concentrations of NAA application slowed down emerging shoot bud in M. afzelii, P. flava, M. erythrophylla and M. philippica. While other species responded positively at some point to increased concentrations of the NAA applications, the P. flava showed retarding effect of phytohormone treatment on its number of leaves. -

Fl. China 19: 218–220. 2011. 53. MITRAGYNA Korthals, Observ. Naucl. Indic. 19. 1839, Nom. Cons., Not Mitragyne R. Brown (1810
Fl. China 19: 218–220. 2011. 53. MITRAGYNA Korthals, Observ. Naucl. Indic. 19. 1839, nom. cons., not Mitragyne R. Brown (1810). 帽蕊木属 mao rui mu shu Chen Tao (陈涛); Charlotte M. Taylor Paradina Pierre ex Pitard; Stephegyne Korthals. Trees, unarmed; buds flattened, with stipules erect and pressed together. Raphides absent. Leaves opposite, sometimes with domatia; stipules caducous, interpetiolar, generally ovate to obovate, sometimes keeled, entire, often well developed. Inflorescences terminal on main stems and axillary branches and often accompanied by reduced, petaloid, and/or bracteate leaves, capitate with globose heads in fascicles, cymes, umbels, or thyrses, sessile to shortly pedunculate, bracteate; bracteoles spatulate to obpyramidal. Flowers sessile, bisexual, monomorphic. Calyx limb truncate to 5-lobed. Corolla cream to yellow-green, funnelform or narrowly salverform, inside glabrous to variously pubescent; lobes 5, valvate in bud. Stamens 5, inserted near corolla throat, exserted or included; filaments short; anthers basifixed, partially to fully exserted. Ovary 2-celled, ovules numerous in each cell on fleshy, pendulous, axile placentas attached in upper third of septum; stigma clavate to mitriform (i.e., upside-down cupular), exserted. Fruit capsular, obovoid to ellipsoid, septicidally then loculicidally dehiscent, cartilaginous to woody, with calyx limb persistent or decidu- ous; seeds numerous, small, somewhat flattened, fusiform to lanceolate, shortly winged at both ends with basal wing sometimes bifid or notched. About seven species: one species in Africa, six species in Asia and Malesia; three species in China. Ridsdale reviewed this genus in detail (Blumea 24: 46–68. 1978) and excluded the African species. H. H. Hsue and H. Wu (in FRPS 71(1): 245. -

Rubiaceae) in Africa and Madagascar
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Springer - Publisher Connector Plant Syst Evol (2010) 285:51–64 DOI 10.1007/s00606-009-0255-8 ORIGINAL ARTICLE Adaptive radiation in Coffea subgenus Coffea L. (Rubiaceae) in Africa and Madagascar Franc¸ois Anthony • Leandro E. C. Diniz • Marie-Christine Combes • Philippe Lashermes Received: 31 July 2009 / Accepted: 28 December 2009 / Published online: 5 March 2010 Ó The Author(s) 2010. This article is published with open access at Springerlink.com Abstract Phylogeographic analysis of the Coffea subge- biogeographic differentiation of coffee species, but they nus Coffea was performed using data on plastid DNA were not congruent with morphological and biochemical sequences and interpreted in relation to biogeographic data classifications, or with the capacity to grow in specific on African rain forest flora. Parsimony and Bayesian analyses environments. Examples of convergent evolution in the of trnL-F, trnT-L and atpB-rbcL intergenic spacers from 24 main clades are given using characters of leaf size, caffeine African species revealed two main clades in the Coffea content and reproductive mode. subgenus Coffea whose distribution overlaps in west equa- torial Africa. Comparison of trnL-F sequences obtained Keywords Africa Á Biogeography Á Coffea Á Evolution Á from GenBank for 45 Coffea species from Cameroon, Phylogeny Á Plastid sequences Á Rubiaceae Madagascar, Grande Comore and the Mascarenes revealed low divergence between African and Madagascan species, suggesting a rapid and radial mode of speciation. A chro- Introduction nological history of the dispersal of the Coffea subgenus Coffea from its centre of origin in Lower Guinea is pro- Coffeeae tribe belongs to the Ixoroideae monophyletic posed. -

(Rubiaceae) D'afrique Tropicale
CONTRIBUTION A L’ÉTUDEBIOLOGIQUE ET TAXONOMIQUE DES MUSSAENDEAE (RUBIACEAE) D’AFRIQUE TROPICALE par Francis’ HALLÉ I. INTRODUCTION La tribu des filussaendeue comprend environ 60 genres, répandus dans toutes les régions intertropicales du globe. Sur ces 60 genres, 4 seule- ment sont véritablement importants du point de vue de la richesse spéci- fique : il s’agit des genres Mussaenda, Sabicea, Urophyllum et Pauridiantha qui comportent respectivement environ 150, 110, 60 et 24 espèces. Tous les autres genres de la tribu ont au maximum 20 espèces, une quarantaine étant même monn- ou bi-spécifiques. L’Afrique Tropicale présente 8 genres de Mussaendeae qui sont : MUSSAENDA . :. ........... 53 espèces en Afrique Tropicale. SABICEA................. 81 - - - PAURIDIANTHA............. 24 - - - STIPULARIA............... 5- -- - PSEUDOMUSSAENDA........ 4- - - ECPOMA.................. genre monospécifique d’Afrique Tropicale. TEMNOPTERYX............ genre monospécifique - - PENTALONCHA............ genre monospécifique 1 - - La présence des trois premiers genres (Mussaenda, Sabicea, Pauri- diaritha) et l’abondance de leur représentation, permet de penser qu’une étude des Mussaendeae peut légitimement être menée sur les seuls repré- sentants africains de cette tribu. LIMITES DE LA TRIBU DES MUSSAENDEAE A la liste donnée ci-dessus nous ajoutons aujourd’hui deux gen- Jes : HEINSIA................... 7 espèces, toutes d’Afrique Tropicale. SACOSPERMA.............. 2 espèces, d’Afrique Tropicale. 1. A cette liste viennent s’ajouter, au moins provisoirement, les genres Pample- fanfha, Sfelecanfha,Commitheca, Poecilocalyx, Rhipidanfha et Empogona : ces petits genres, tous mono- ou bispécifiques, résultent du demembrement du genre Urophyllum par Bremekamp (1941) et leurs affinitks sont encore imprkcises. O. E”,. S. T. O. M. - 267 - Le genre Heinsia, placé par Hutchinson et Dalziel dans la tribu hétérogène des Hamelieae, avait déjà été rapproché du geme Mussaenda par B. -
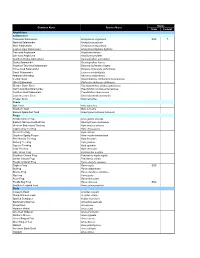
St. Joseph Bay Native Species List
Status Common Name Species Name State Federal Amphibians Salamanders Flatwoods Salamander Ambystoma cingulatum SSC T Marbled Salamander Ambystoma opacum Mole Salamander Ambystoma talpoideum Eastern Tiger Salamander Ambystoma tigrinum tigrinum Two-toed Amphiuma Amphiuma means One-toed Amphiuma Amphiuma pholeter Southern Dusky Salamander Desmognathus auriculatus Dusky Salamander Desmognathus fuscus Southern Two-lined Salamander Eurycea bislineata cirrigera Three-lined Salamander Eurycea longicauda guttolineata Dwarf Salamander Eurycea quadridigitata Alabama Waterdog Necturus alabamensis Central Newt Notophthalmus viridescens louisianensis Slimy Salamander Plethodon glutinosus glutinosus Slender Dwarf Siren Pseudobranchus striatus spheniscus Gulf Coast Mud Salamander Pseudotriton montanus flavissimus Southern Red Salamander Pseudotriton ruber vioscai Eastern Lesser Siren Siren intermedia intermedia Greater Siren Siren lacertina Toads Oak Toad Bufo quercicus Southern Toad Bufo terrestris Eastern Spadefoot Toad Scaphiopus holbrooki holbrooki Frogs Florida Cricket Frog Acris gryllus dorsalis Eastern Narrow-mouthed Frog Gastrophryne carolinensis Western Bird-voiced Treefrog Hyla avivoca avivoca Cope's Gray Treefrog Hyla chrysoscelis Green Treefrog Hyla cinerea Southern Spring Peeper Hyla crucifer bartramiana Pine Woods Treefrog Hyla femoralis Barking Treefrog Hyla gratiosa Squirrel Treefrog Hyla squirella Gray Treefrog Hyla versicolor Little Grass Frog Limnaoedus ocularis Southern Chorus Frog Pseudacris nigrita nigrita Ornate Chorus Frog Pseudacris -

Pollen Morphology of Some Cuban Guettarda Species
This article was downloaded by: [200.196.73.69] On: 27 April 2013, At: 13:36 Publisher: Taylor & Francis Informa Ltd Registered in England and Wales Registered Number: 1072954 Registered office: Mortimer House, 37-41 Mortimer Street, London W1T 3JH, UK Grana Publication details, including instructions for authors and subscription information: http://www.tandfonline.com/loi/sgra20 Pollen morphology of some Cuban Guettarda species (Rubiaceae: Guettardeae) Lázara Sotolongo Molina , Maira Fernández Zequeira & Pedro Herrera Oliver Version of record first published: 05 Nov 2010. To cite this article: Lázara Sotolongo Molina , Maira Fernández Zequeira & Pedro Herrera Oliver (2002): Pollen morphology of some Cuban Guettarda species (Rubiaceae: Guettardeae), Grana, 41:3, 142-148 To link to this article: http://dx.doi.org/10.1080/001731302321042605 PLEASE SCROLL DOWN FOR ARTICLE Full terms and conditions of use: http://www.tandfonline.com/page/terms-and-conditions This article may be used for research, teaching, and private study purposes. Any substantial or systematic reproduction, redistribution, reselling, loan, sub-licensing, systematic supply, or distribution in any form to anyone is expressly forbidden. The publisher does not give any warranty express or implied or make any representation that the contents will be complete or accurate or up to date. The accuracy of any instructions, formulae, and drug doses should be independently verified with primary sources. The publisher shall not be liable for any loss, actions, claims, proceedings, demand, or costs or damages whatsoever or howsoever caused arising directly or indirectly in connection with or arising out of the use of this material. Grana 41: 142–148, 2002 Pollen morphology of some Cuban Guettarda species (Rubiaceae: Guettardeae) LA´ ZARA SOTOLONGO MOLINA, MAIRA FERNA´ NDEZ ZEQUEIRA and PEDRO HERRERA OLIVER Sotolongo Molina, L., Ferna´ndez Zequeira, M. -

Tree and Tree-Like Species of Mexico: Asteraceae, Leguminosae, and Rubiaceae
Revista Mexicana de Biodiversidad 84: 439-470, 2013 Revista Mexicana de Biodiversidad 84: 439-470, 2013 DOI: 10.7550/rmb.32013 DOI: 10.7550/rmb.32013439 Tree and tree-like species of Mexico: Asteraceae, Leguminosae, and Rubiaceae Especies arbóreas y arborescentes de México: Asteraceae, Leguminosae y Rubiaceae Martin Ricker , Héctor M. Hernández, Mario Sousa and Helga Ochoterena Herbario Nacional de México, Departamento de Botánica, Instituto de Biología, Universidad Nacional Autónoma de México. Apartado postal 70- 233, 04510 México D. F., Mexico. [email protected] Abstract. Trees or tree-like plants are defined here broadly as perennial, self-supporting plants with a total height of at least 5 m (without ascending leaves or inflorescences), and with one or several erect stems with a diameter of at least 10 cm. We continue our compilation of an updated list of all native Mexican tree species with the dicotyledonous families Asteraceae (36 species, 39% endemic), Leguminosae with its 3 subfamilies (449 species, 41% endemic), and Rubiaceae (134 species, 24% endemic). The tallest tree species reach 20 m in the Asteraceae, 70 m in the Leguminosae, and also 70 m in the Rubiaceae. The species-richest genus is Lonchocarpus with 67 tree species in Mexico. Three legume genera are endemic to Mexico (Conzattia, Hesperothamnus, and Heteroflorum). The appendix lists all species, including their original publication, references of taxonomic revisions, existence of subspecies or varieties, maximum height in Mexico, and endemism status. Key words: biodiversity, flora, tree definition. Resumen. Las plantas arbóreas o arborescentes se definen aquí en un sentido amplio como plantas perennes que se pueden sostener por sí solas, con una altura total de al menos 5 m (sin considerar hojas o inflorescencias ascendentes) y con uno o varios tallos erectos de un diámetro de al menos 10 cm. -

Differentiation of Mitragyna Speciosa, a Narcotic Plant, from Allied Mitragyna Species Using DNA Barcoding-High-Resolution Melting (Bar-HRM) Analysis
Differentiation of Mitragyna Speciosa, a Narcotic Plant, From Allied Mitragyna Species Using DNA Barcoding-high-resolution Melting (Bar-HRM) Analysis Chayapol Tungphatthong Chulalongkorn University Santhosh Kumar J Urumarudappa Chulalongkorn University Supita Awachai Chulalongkorn University Thongchai Sooksawate Chulalongkorn University Suchada Sukrong ( [email protected] ) Chulalongkorn University Research Article Keywords: Mitragyna speciosa, Kratom, Rubiaceae, Bar-HRM, Narcotic plant Posted Date: November 25th, 2020 DOI: https://doi.org/10.21203/rs.3.rs-109985/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Version of Record: A version of this preprint was published at Scientic Reports on March 24th, 2021. See the published version at https://doi.org/10.1038/s41598-021-86228-9. Page 1/16 Abstract Mitragyna speciosa (Korth.) Havil. [MS], or “kratom” in Thai, is the only narcotic species among the four species of Mitragyna in Thailand, which also include Mitragyna diversifolia (Wall. ex G. Don) Havil. [MD], Mitragyna hirsuta Havil. [MH], and Mitragyna rotundifolia (Roxb.) O. Kuntze [MR]. M. speciosa is a tropical tree belonging to the Rubiaceae family and has been prohibited by law in Thailand. However, it has been extensively covered in national and international news, as its abuse has become more popular. M. speciosa is a narcotic plant and has been used as an opium substitute and traditionally used for the treatment of chronic pain and various illnesses. Due to morphological disparities in the genus, the identication of plants in various forms, including fresh leaves, dried leaf powder, and nished products, is dicult. In this study, DNA barcoding combined with high-resolution melting (Bar-HRM) analysis was performed to differentiate M.