Mutation Causing Von Willebrand's Disease in Scottish Terriers
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Developing a Better Breeding Program
Pedigree Analysis and How Breeding Decisions Affect Genes Jerold S Bell DVM, Clinical Associate Professor of Genetics, Tufts Cummings School of Veterinary Medicine To some breeders, determining which traits will appear in may be phenotypically uniform, but will rarely breed true due the offspring of a mating is like rolling the dice - a to the mix of dissimilar genes. combination of luck and chance. For others, producing certain traits involves more skill than luck - the result of One reason to outbreed would be to bring in new traits that careful study and planning. As breeders, you must your breeding stock does not possess. While the parents may understand how matings manipulate genes within your be genetically dissimilar, you should choose a mate that breeding stock to produce the kinds of offspring you desire. corrects your breeding animal's faults but complements its good traits. It is not unusual to produce an excellent quality When evaluating your breeding program, remember that individual from an outbred litter. The abundance of genetic most traits you're seeking cannot be changed, fixed or variability can place all the right pieces in one individual. created in a single generation. The more information you Many top-winning show animals are outbred. Consequently, can obtain on how certain traits have been transmitted by however, they may have low inbreeding coefficients and may your animal's ancestors, the better you can prioritize your lack the ability to uniformly pass on their good traits to their breeding goals. Tens of thousands of genes interact to offspring. After an outbreeding, breeders may want to breed produce a single individual. -

Frequently Asked Questions
APPLYING SHAMPOO & CONDITIONER For best results apply the shampoo & conditioner in the following order: 1. belly & Determine the appropriate shampoo & conditioner for your breed. chest 2. legs and feet 3. neck & face 4. back SHORT COATS need LEMON, MEDIUM COATS need BANANA and bottom. Let soak for 5 full minutes. These LONG COATS need GREEN APPLE. are the sebaceous areas where most dirt and toxins enter the pet’s system. SHAMPOO: Dilute 1 part shampoo to 3 parts water in a clean container. Wet the coat & apply shampoo. Soak for 5 full minutes. Rinse. CONDITIONER: Dilute 1 part conditioner to 3 parts water in a clean container. Apply conditioner to coat. Soak for 5 full minutes. Rinse. Don’t see your breed? Please visit Please visit our info site http://isbusa.com/ instructions/akc-breeds-by-coat-type/ Frequently Asked Questions: SHORT COATS MEDIUM COATS LONG COATS Why are there different products for different American Foxhound Bernese Mountain Dog Afghan Hound breeds? All pet coats need oils, minerals and Staffordshire Terrier Brussels Griffon Bearded Collie collagen to thrive. However short coats need Basenji Cairn Terrier Bedlington Terrier more oils, medium coats need more minerals Basset Hound Cavalier King Charles Bichon Frise and long coats need more collagen. These Beagle Chesapeake Bay Retriever Bolonese products are created to provide the right Beauceron Chow Chow Chinese Crested balance of each for all coat types. Bloodhound Cocker Spaniel Coton De Tulear Why do I have to bathe per your instructions? Boston Terrier German Shepherd Havanese Since pets absorb dirt & toxins from the ground Boxer Golden Retriever Japanese Chin up into their sebaceous areas (tummy, feet, Great Dane Irish Wolfhound Lhasa Apso legs and other areas with less hair) it is Bull Terrier Keeshond Longhaired Dachshund important that the products are applied there Chihuahua Labrador Retriever Maltese first & allowed to soak for 5 min. -

Pedigree Analysis and Optimisation of the Breeding Programme of the Markiesje and the Stabyhoun
Pedigree analysis and optimisation of the breeding programme of the Markiesje and the Stabyhoun Aiming to improve health and welfare and maintain genetic diversity Harmen Doekes REG.NR.: 920809186050 Major thesis Animal Breeding and Genetics (ABG-80436) January, 2016 Supervisors/examiners: Piter Bijma - Animal Breeding and Genomics Centre, Wageningen University Kor Oldenbroek - Centre for Genetic Resources the Netherlands (CGN), Wageningen UR Jack Windig - Animal Breeding & Genomics Centre, Wageningen UR Livestock Research Thesis: Animal Breeding and Genomics Centre Pedigree analysis and optimisation of the breeding programme of the Markiesje and the Stabyhoun Aiming to improve health and welfare and maintain genetic diversity Harmen Doekes REG.NR.: 920809186050 Major thesis Animal Breeding and Genetics (ABG-80436) January, 2016 Supervisors: Kor Oldenbroek - Centre for Genetic Resources the Netherlands (CGN), Wageningen UR Jack Windig - Animal Breeding & Genomics Centre, Wageningen UR Livestock Research Examiners: Piter Bijma - Animal Breeding and Genomics Centre, Wageningen University Kor Oldenbroek - Centre for Genetic Resources the Netherlands (CGN), Wageningen UR Commissioned by: Nederlandse Markiesjes Vereniging Nederlandse Vereniging voor Stabij- en Wetterhounen Preface This major thesis is submitted in partial fulfilment of the requirements for the degree of Master of Animal Sciences of Wageningen University, the Netherlands. It comprises an unpublished study on the genetic status of two Dutch dog breeds, the Markiesje and the Stabyhoun. that was commissioned by the Breed Clubs of the breeds, the ‘Nederlandse Markiesjes Vereniging’ and the ‘Nederlandse Vereniging voor Stabij- en Wetterhounen’. It was written for readers with limited pre-knowledge. Although the thesis focusses on two breeds, it addresses issues that are found in many dog breeds. -

The Relation Between Canine Hip Dysplasia, Genetic Diversity and Inbreeding by Breed
Open Journal of Veterinary Medicine, 2014, 4, 67-71 Published Online May 2014 in SciRes. http://www.scirp.org/journal/ojvm http://dx.doi.org/10.4236/ojvm.2014.45008 The Relation between Canine Hip Dysplasia, Genetic Diversity and Inbreeding by Breed Frank H. Comhaire Department of Internal Medicine, Ghent University, Ghent, Belgium Email: [email protected] Received 18 March 2014; revised 18 April 2014; accepted 28 April 2014 Copyright © 2014 by author and Scientific Research Publishing Inc. This work is licensed under the Creative Commons Attribution International License (CC BY). http://creativecommons.org/licenses/by/4.0/ Abstract Objectives: To assess the relation between the prevalence of canine hip dysplasia, inbreeding and genetic diversity by breed. Methods: Retrospective pedigree analysis of 9 breeds based on a ref- erence population of 41,728 individuals, and hip dysplasia assessment in 1745 dogs. Results: Hip dysplasia was less common among breeds with higher coefficient of inbreeding, lower genetic di- versity, and highest contribution of one single ancestor to the population. Inbreeding not exceed- ing 3.25% should be considered safe since it will maintain a sufficiently high genetic diversity within the breed. Clinical Significance: Together with published data on single breeds, the present findings question the general assumption that line-breeding or in-breeding has an adverse effect on the prevalence of hip dysplasia. Hip assessment is indicated in all breeds, but better methods are needed for selecting dogs suitable for reproduction. Keywords Genetic Diversity, Effective Population Size, Inbreeding, Hip Dysplasia 1. Introduction A high degree of inbreeding increases the probability of homozygosity of recessive genes, and enhances the risk of hereditary diseases coming to expression. -

Ranked by Temperament
Comparing Temperament and Breed temperament was determined using the American 114 DOG BREEDS Popularity in Dog Breeds in Temperament Test Society's (ATTS) cumulative test RANKED BY TEMPERAMENT the United States result data since 1977, and breed popularity was determined using the American Kennel Club's (AKC) 2018 ranking based on total breed registrations. Number Tested <201 201-400 401-600 601-800 801-1000 >1000 American Kennel Club 50% 60% 70% 80% 90% 1. Labrador 100% Popularity Passed 2. German Retriever Passed Shepherd 3. Mixed Breed 7. Beagle Dog 4. Golden Retriever More Popular 8. Poodle 11. Rottweiler 5. French Bulldog 6. Bulldog (Miniature)10. Poodle (Toy) 15. Dachshund (all varieties) 9. Poodle (Standard) 17. Siberian 16. Pembroke 13. Yorkshire 14. Boxer 18. Australian Terrier Husky Welsh Corgi Shepherd More Popular 12. German Shorthaired 21. Cavalier King Pointer Charles Spaniel 29. English 28. Brittany 20. Doberman Spaniel 22. Miniature Pinscher 19. Great Dane Springer Spaniel 24. Boston 27. Shetland Schnauzer Terrier Sheepdog NOTE: We excluded breeds that had fewer 25. Bernese 30. Pug Mountain Dog 33. English than 30 individual dogs tested. 23. Shih Tzu 38. Weimaraner 32. Cocker 35. Cane Corso Cocker Spaniel Spaniel 26. Pomeranian 31. Mastiff 36. Chihuahua 34. Vizsla 40. Basset Hound 37. Border Collie 41. Newfoundland 46. Bichon 39. Collie Frise 42. Rhodesian 44. Belgian 47. Akita Ridgeback Malinois 49. Bloodhound 48. Saint Bernard 45. Chesapeake 51. Bullmastiff Bay Retriever 43. West Highland White Terrier 50. Portuguese 54. Australian Water Dog Cattle Dog 56. Scottish 53. Papillon Terrier 52. Soft Coated 55. Dalmatian Wheaten Terrier 57. -

Review of the Current State of Genetic Testing - a Living Resource
Review of the Current State of Genetic Testing - A Living Resource Prepared by Liza Gershony, DVM, PhD and Anita Oberbauer, PhD of the University of California, Davis Editorial input by Leigh Anne Clark, PhD of Clemson University July, 2020 Contents Introduction .................................................................................................................................................. 1 I. The Basics ......................................................................................................................................... 2 II. Modes of Inheritance ....................................................................................................................... 7 a. Mendelian Inheritance and Punnett Squares ................................................................................. 7 b. Non-Mendelian Inheritance ........................................................................................................... 10 III. Genetic Selection and Populations ................................................................................................ 13 IV. Dog Breeds as Populations ............................................................................................................. 15 V. Canine Genetic Tests ...................................................................................................................... 16 a. Direct and Indirect Tests ................................................................................................................ 17 b. Single -
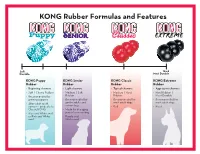
Kong Size Chart
KONG Rubber Formulas and Features So Hard Durable Most Durable KONG Puppy KONG Senior KONG Classic KONG Extreme Rubber Rubber Rubber Rubber • Beginning chewers • Light chewers • Typical chewers • Aggressive chewers • Soft / Chewy Rubber • Medium / Soft • Medium / Hard • Hard Rubber / • Recommended for Rubber Rubber Most Durable growing puppies • Recommended for • Recommended for • Recommended for • After adult teeth gentle adults and most adult dogs most adult dogs come in - graduate to senior dogs • Red • Black Classic KONG • Made for the aging • Blue and White swirl jaws of a senior dog or Pink and White • Purple and swirl White swirl Recommended Sizing Always supervise new toy use. Dog Breeds Sizing Sizing Sizing Sizing Dog Breeds Sizing Sizing Sizing Sizing Akita L* XL, XXL XL, XXL SEE BELOW*** Lhasa Apso S, M S**, M S, M M Alaskan Malamute L* XL, XXL XL, XXL SEE BELOW*** Maltese S S S S Basset Hound M, L L, XL L, XL L Mastiff L* XXL XXL SEE BELOW*** Beagle S, M M, L M, L M, L Pit Bull/Am. Stat. L* XL**, XXL** XL**, XXL SEE BELOW*** Bichon S, M S**, M S, M M Pointer M*, L L, XL L, XL L Boston Terrier S, M M, L M, L M, L Pomeranian S XS, S S S Boxer L L**, XL L, XL L Poodle (std.) M*, L L, XL L, XL L Brittany M, L M, L M, L M, L Poodle (min) S, M S, M S, M M Bulldog L XL**, XXL** XL, XXL SEE BELOW*** Pug S, M M, L M, L M, L Chihuahua S XS, S S S Rhodesian Ridgeback M*, L L**, XL, XXL L, XL, XXL L Chow Chow L L**, XL L, XL L Rottweiler L* XL**, XXL XL**, XXL SEE BELOW*** Cocker Spaniel S, M M, L M, L M, L Saint Bernard L* XXL XXL SEE -
Draft for a Discussion on Basic Genetics.Pdf
2 Foreword. “Mens sana in corpore sano” : a healthy mind in a healthy body. The health of the Rottweiler, worldwide, lies close to our heart and is embedded in the Constitution of the International Federation of Rottweilerfriends (IFR). It’s reference to the health of all breeding dogs, implies that the Federation and its Memberclubs must pay particular attention to genetic disorders. For the Rottweiler, Hip Dysplasia (HD) and Elbow Dysplasia (ED) were the first disorders that worldwide led to regulations meant to diminish the genetic affection of the breed. The measures that were taken by breed clubs and/or kennelclubs, mostly by excluding the dogs that are affected by the disease from breeding, have proven to be quite effective, although not enough to completely eliminate the genetic factor and remove it from the genepool Still they have helped to limit the number of affected dogs and to avoid a lot of suffering. In this brochure you will find a brief review of just a few of the genetic diseases that are said to have entered the Rottweiler’s genepool. 1 The list is not new, nor the particular diseases on it. Even the way of approaching the problem is known for a long time and was amongst others confirmed in the International Breeding Strategies of the FCI that were approved in 2009. Indeed, not just for the Rottweiler but for many other breeds too, cynology has sounded a loud alarm concerning an ever more reduced genetic diversity among certain canine breeds, causing not only extreme phenotypes (2) but also physical and health problems. -

Scottish Terrier Club of America Records Finding Aid Prepared by Kari Dalane, 2009; Additions, Edits, and Conversion of Legacy Finding Aid by Brynn White, 2016
Scottish Terrier Club of America records Finding aid prepared by Kari Dalane, 2009; Additions, edits, and conversion of legacy finding aid by Brynn White, 2016 This finding aid was produced using the Archivists' Toolkit May 23, 2016 Describing Archives: A Content Standard American Kennel Club Archives Scottish Terrier Club of America records Table of Contents Summary Information ................................................................................................................................. 3 Historical Information....................................................................................................................................4 Scope and Contents Note.............................................................................................................................. 6 Arrangement...................................................................................................................................................8 Administrative Information .........................................................................................................................8 Collection Inventory...................................................................................................................................... 9 I. Club Administration.............................................................................................................................9 II. Publications...................................................................................................................................... -

An Insight Into Population Structure and Gene Flow Within Purebred Cats
J. Anim. Breed. Genet. ISSN 0931-2668 ORIGINAL ARTICLE An insight into population structure and gene flow within pure-bred cats G. Leroy1,2, E. Vernet3, M.B. Pautet3 & X. Rognon1,2 1 UMR1313 Gen etique Animale et Biologie Integrative, AgroParisTech, Paris, France 2 UMR1313 Gen etique Animale et Biologie Integrative, INRA, Jouy-en-Josas, France 3 LOOF, Pantin, France Keywords Summary Cat; gene flow; genetic variability; inbreeding; Investigation of genetic structure on the basis of pedigree information population structure. requires indicators adapted to the specific context of the populations stud- Correspondence ied. On the basis of pedigree-based estimates of diversity, we analysed G. Leroy, UMR1313 Gen etique Animale et genetic diversity, mating practices and gene flow among eight cat popula- Biologie Integrative, AgroParisTech, 16 rue tions raised in France, five of them being single breeds and three consist- Claude Bernard, F-75231 Paris 05, France. ing of breed groups with varieties that may interbreed. When computed Tel: +33 (0) 1 44 08 17 46; Fax: +33 (0) 1 44 08 on the basis of coancestry rate, effective population sizes ranged from 127 86 22; E-mail: [email protected] to 1406, while the contribution of founders from other breeds ranged from 0.7 to 16.4%. In the five breeds, FIS ranged between 0.96 and 1.83%, with Received: 29 October 2012; this result being related to mating practices such as close inbreeding (on accepted: 27 April 2013 average 5% of individuals being inbred within two generations). Within the three groups of varieties studied, FIT ranged from 1.59 to 3%, while FST values were estimated between 0.04 and 0.91%, which was linked to various amounts of gene exchanges between subpopulations at the paren- tal level. -

Baskerville Ultra Muzzle Breed Guide. Sizes Are Available in 1 - 6 and Are for Typical Adult Dogs & Bitches
Baskerville Ultra Muzzle Breed Guide. Sizes are available in 1 - 6 and are for typical adult dogs & bitches. Juveniles may need a size smaller. ‡ = not recommended. The number next to the breeds below is the recommended size. Boston Terrier ‡ Bulldog ‡ King Charles Spaniel ‡ Lhasa Apso ‡ Pekingese ‡ Pug ‡ St Bernard ‡ Shih Tzu ‡ Afghan Hound 5 Airedale 5 Alaskan Malamute 5 American Cocker 2 American Staffordshire 6 Australian Cattle Dog 3 Australian Shepherd 3 Basenji 2 Basset Hound 5 Beagle 3 Bearded Collie 3 Bedlington Terrier 2 Belgian Shepherd 5 Bernese MD 5 Bichon Frisé 1 Border Collie 3 Border Terrier 2 Borzoi 5 Bouvier 6 Boxer 6 Briard 5 Brittany Spaniel 5 Buhund 2 Bull Mastiff 6 Bull Terrier 5 Cairn Terrier 2 Cavalier Spaniel 2 Chow Chow 5 Chesapeake Bay Retriever 5 Cocker (English) 3 Corgi 3 Dachshund Miniature 1 Dachshund Standard 1 Dalmatian 4 Dobermann 5 Elkhound 4 English Setter 5 Flat Coated Retriever 5 Foxhound 5 Fox Terrier 2 German Shepherd 5 Golden Retriever 5 Gordon Setter 5 Great Dane 6 Greyhound 5 Hungarian Vizsla 3 Irish Setter 5 Irish Water Spaniel 3 Irish Wolfhound 6 Jack Russell 2 Japanese Akita 6 Keeshond 3 Kerry Blue Terrier 4 Labrador Retriever 5 Lakeland Terrier 2 Lurcher 5 Maltese Terrier 1 Maremma Sheepdog 5 Mastiff 6 Munsterlander 5 Newfoundland 6 Norfolk/Norwich Terrier 1 Old English Sheepdog 5 Papillon N/A Pharaoh Hound 5 Pit Bull 6 Pointers 4 Poodle Toy 1 Poodle Standard 3 Pyrenean MD 6 Ridgeback 5 Rottweiler 6 Rough Collie 3 Saluki 3 Samoyed 4 Schnauzer Miniature 2 Schnauzer 3 Schnauzer Giant 6 Scottish Terrier 3 Sheltie 2 Shiba Inu 2 Siberian Husky 5 Soft Coated Wheaten 4 Springer Spaniel 4 Staff Bull Terrier 6 Weimaraner 5 Welsh Terrier 3 West Highland White 2 Whippet 2 Yorkshire Terrier 1 . -

A Short History of the Scottish Terrier Breed Standard by Cindy Cooke
A Short History of the Scottish Terrier Breed Standard Article written by Cindy Cooke The Scottish Terrier is often described as the oldest breed in Scotland, but no one can accurately determine exactly how the breed evolved. Few outsiders were interested enough in the highland dogs to research and write about them until the late 1870s when Capt. W. W. Mackie spent several winters touring Scotland to see Scottish Terriers in their native habitat. Mackie, who described himself as having “terriers on the brain,” kept a diary in which he described the many types of dogs that were all described as “Scottish” terriers. According to Mackie’s diary, every highland community boasted a gamekeeper or a deer forester who kept a pack of terriers to rid the community of foxes, otters, weasels, stoats, rats and other vermin. The dogs varied widely in size, color and ear carriage, but every terrier man in the highlands of Scotland thought his own strain of dogs was the best and the truest Scottish Terrier.[1] Mackie already had a mental picture of the correct type of Scottish Terrier: They have a thoroughly Scottish look about them, and would shame two-thirds of the “messans” we sometimes see on the show-bench. They are of divers colours, from a grizzly brindle to sandy; in weight they will run from 17 to 20 lbs; knowing-looking big heads, sharp muzzles, powerful jaws, very large teeth, ears semi-erect, or “cock-and-a-half cock”; stout bony legs, the fore ones slightly bent. If these little dogs have any faults, I would say the fault lay in their ears being heavy, and their tails being inclined to curl slightly.