Data Engineering for HPC with Python
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Apache Flink™: Stream and Batch Processing in a Single Engine
Apache Flink™: Stream and Batch Processing in a Single Engine Paris Carboney Stephan Ewenz Seif Haridiy Asterios Katsifodimos* Volker Markl* Kostas Tzoumasz yKTH & SICS Sweden zdata Artisans *TU Berlin & DFKI parisc,[email protected] fi[email protected] fi[email protected] Abstract Apache Flink1 is an open-source system for processing streaming and batch data. Flink is built on the philosophy that many classes of data processing applications, including real-time analytics, continu- ous data pipelines, historic data processing (batch), and iterative algorithms (machine learning, graph analysis) can be expressed and executed as pipelined fault-tolerant dataflows. In this paper, we present Flink’s architecture and expand on how a (seemingly diverse) set of use cases can be unified under a single execution model. 1 Introduction Data-stream processing (e.g., as exemplified by complex event processing systems) and static (batch) data pro- cessing (e.g., as exemplified by MPP databases and Hadoop) were traditionally considered as two very different types of applications. They were programmed using different programming models and APIs, and were exe- cuted by different systems (e.g., dedicated streaming systems such as Apache Storm, IBM Infosphere Streams, Microsoft StreamInsight, or Streambase versus relational databases or execution engines for Hadoop, including Apache Spark and Apache Drill). Traditionally, batch data analysis made up for the lion’s share of the use cases, data sizes, and market, while streaming data analysis mostly served specialized applications. It is becoming more and more apparent, however, that a huge number of today’s large-scale data processing use cases handle data that is, in reality, produced continuously over time. -

Mastering Machine Learning with Scikit-Learn
www.it-ebooks.info Mastering Machine Learning with scikit-learn Apply effective learning algorithms to real-world problems using scikit-learn Gavin Hackeling BIRMINGHAM - MUMBAI www.it-ebooks.info Mastering Machine Learning with scikit-learn Copyright © 2014 Packt Publishing All rights reserved. No part of this book may be reproduced, stored in a retrieval system, or transmitted in any form or by any means, without the prior written permission of the publisher, except in the case of brief quotations embedded in critical articles or reviews. Every effort has been made in the preparation of this book to ensure the accuracy of the information presented. However, the information contained in this book is sold without warranty, either express or implied. Neither the author, nor Packt Publishing, and its dealers and distributors will be held liable for any damages caused or alleged to be caused directly or indirectly by this book. Packt Publishing has endeavored to provide trademark information about all of the companies and products mentioned in this book by the appropriate use of capitals. However, Packt Publishing cannot guarantee the accuracy of this information. First published: October 2014 Production reference: 1221014 Published by Packt Publishing Ltd. Livery Place 35 Livery Street Birmingham B3 2PB, UK. ISBN 978-1-78398-836-5 www.packtpub.com Cover image by Amy-Lee Winfield [email protected]( ) www.it-ebooks.info Credits Author Project Coordinator Gavin Hackeling Danuta Jones Reviewers Proofreaders Fahad Arshad Simran Bhogal Sarah -

Installing Python Ø Basics of Programming • Data Types • Functions • List Ø Numpy Library Ø Pandas Library What Is Python?
Python in Data Science Programming Ha Khanh Nguyen Agenda Ø What is Python? Ø The Python Ecosystem • Installing Python Ø Basics of Programming • Data Types • Functions • List Ø NumPy Library Ø Pandas Library What is Python? • “Python is an interpreted high-level general-purpose programming language.” – Wikipedia • It supports multiple programming paradigms, including: • Structured/procedural • Object-oriented • Functional The Python Ecosystem Source: Fabien Maussion’s “Getting started with Python” Workshop Installing Python • Install Python through Anaconda/Miniconda. • This allows you to create and work in Python environments. • You can create multiple environments as needed. • Highly recommended: install Python through Miniconda. • Are we ready to “play” with Python yet? • Almost! • Most data scientists use Python through Jupyter Notebook, a web application that allows you to create a virtual notebook containing both text and code! • Python Installation tutorial: [Mac OS X] [Windows] For today’s seminar • Due to the time limitation, we will be using Google Colab instead. • Google Colab is a free Jupyter notebook environment that runs entirely in the cloud, so you can run Python code, write report in Jupyter Notebook even without installing the Python ecosystem. • It is NOT a complete alternative to installing Python on your local computer. • But it is a quick and easy way to get started/try out Python. • You will need to log in using your university account (possible for some schools) or your personal Google account. Basics of Programming Indentations – Not Braces • Other languages (R, C++, Java, etc.) use braces to structure code. # R code (not Python) a = 10 if (a < 5) { result = TRUE b = 0 } else { result = FALSE b = 100 } a = 10 if (a < 5) { result = TRUE; b = 0} else {result = FALSE; b = 100} Basics of Programming Indentations – Not Braces • Python uses whitespaces (tabs or spaces) to structure code instead. -

Quick Install for AWS EMR
Quick Install for AWS EMR Version: 6.8 Doc Build Date: 01/21/2020 Copyright © Trifacta Inc. 2020 - All Rights Reserved. CONFIDENTIAL These materials (the “Documentation”) are the confidential and proprietary information of Trifacta Inc. and may not be reproduced, modified, or distributed without the prior written permission of Trifacta Inc. EXCEPT AS OTHERWISE PROVIDED IN AN EXPRESS WRITTEN AGREEMENT, TRIFACTA INC. PROVIDES THIS DOCUMENTATION AS-IS AND WITHOUT WARRANTY AND TRIFACTA INC. DISCLAIMS ALL EXPRESS AND IMPLIED WARRANTIES TO THE EXTENT PERMITTED, INCLUDING WITHOUT LIMITATION THE IMPLIED WARRANTIES OF MERCHANTABILITY, NON-INFRINGEMENT AND FITNESS FOR A PARTICULAR PURPOSE AND UNDER NO CIRCUMSTANCES WILL TRIFACTA INC. BE LIABLE FOR ANY AMOUNT GREATER THAN ONE HUNDRED DOLLARS ($100) BASED ON ANY USE OF THE DOCUMENTATION. For third-party license information, please select About Trifacta from the Help menu. 1. Release Notes . 4 1.1 Changes to System Behavior . 4 1.1.1 Changes to the Language . 4 1.1.2 Changes to the APIs . 18 1.1.3 Changes to Configuration 23 1.1.4 Changes to the Object Model . 26 1.2 Release Notes 6.8 . 30 1.3 Release Notes 6.4 . 36 1.4 Release Notes 6.0 . 42 1.5 Release Notes 5.1 . 49 2. Quick Start 55 2.1 Install from AWS Marketplace with EMR . 55 2.2 Upgrade for AWS Marketplace with EMR . 62 3. Configure 62 3.1 Configure for AWS . 62 3.1.1 Configure for EC2 Role-Based Authentication . 68 3.1.2 Enable S3 Access . 70 3.1.2.1 Create Redshift Connections 81 3.1.3 Configure for EMR . -

Evaluation of SPARQL Queries on Apache Flink
applied sciences Article SPARQL2Flink: Evaluation of SPARQL Queries on Apache Flink Oscar Ceballos 1 , Carlos Alberto Ramírez Restrepo 2 , María Constanza Pabón 2 , Andres M. Castillo 1,* and Oscar Corcho 3 1 Escuela de Ingeniería de Sistemas y Computación, Universidad del Valle, Ciudad Universitaria Meléndez Calle 13 No. 100-00, Cali 760032, Colombia; [email protected] 2 Departamento de Electrónica y Ciencias de la Computación, Pontificia Universidad Javeriana Cali, Calle 18 No. 118-250, Cali 760031, Colombia; [email protected] (C.A.R.R.); [email protected] (M.C.P.) 3 Ontology Engineering Group, Universidad Politécnica de Madrid, Campus de Montegancedo, Boadilla del Monte, 28660 Madrid, Spain; ocorcho@fi.upm.es * Correspondence: [email protected] Abstract: Existing SPARQL query engines and triple stores are continuously improved to handle more massive datasets. Several approaches have been developed in this context proposing the storage and querying of RDF data in a distributed fashion, mainly using the MapReduce Programming Model and Hadoop-based ecosystems. New trends in Big Data technologies have also emerged (e.g., Apache Spark, Apache Flink); they use distributed in-memory processing and promise to deliver higher data processing performance. In this paper, we present a formal interpretation of some PACT transformations implemented in the Apache Flink DataSet API. We use this formalization to provide a mapping to translate a SPARQL query to a Flink program. The mapping was implemented in a prototype used to determine the correctness and performance of the solution. The source code of the Citation: Ceballos, O.; Ramírez project is available in Github under the MIT license. -

Delft University of Technology Arrowsam In-Memory Genomics
Delft University of Technology ArrowSAM In-Memory Genomics Data Processing Using Apache Arrow Ahmad, Tanveer; Ahmed, Nauman; Peltenburg, Johan; Al-Ars, Zaid DOI 10.1109/ICCAIS48893.2020.9096725 Publication date 2020 Document Version Accepted author manuscript Published in 2020 3rd International Conference on Computer Applications & Information Security (ICCAIS) Citation (APA) Ahmad, T., Ahmed, N., Peltenburg, J., & Al-Ars, Z. (2020). ArrowSAM: In-Memory Genomics Data Processing Using Apache Arrow. In 2020 3rd International Conference on Computer Applications & Information Security (ICCAIS): Proceedings (pp. 1-6). [9096725] IEEE . https://doi.org/10.1109/ICCAIS48893.2020.9096725 Important note To cite this publication, please use the final published version (if applicable). Please check the document version above. Copyright Other than for strictly personal use, it is not permitted to download, forward or distribute the text or part of it, without the consent of the author(s) and/or copyright holder(s), unless the work is under an open content license such as Creative Commons. Takedown policy Please contact us and provide details if you believe this document breaches copyrights. We will remove access to the work immediately and investigate your claim. This work is downloaded from Delft University of Technology. For technical reasons the number of authors shown on this cover page is limited to a maximum of 10. © 2020 IEEE. Personal use of this material is permitted. Permission from IEEE must be obtained for all other uses, in any current or future media, including reprinting/republishing this material for advertising or promotional purposes, creating new collective works, for resale or redistribution to servers or lists, or reuse of any copyrighted component of this work in other works. -

The Platform Inside and out Release 0.8
The Platform Inside and Out Release 0.8 Joshua Patterson – GM, Data Science RAPIDS End-to-End Accelerated GPU Data Science Data Preparation Model Training Visualization Dask cuDF cuIO cuML cuGraph PyTorch Chainer MxNet cuXfilter <> pyViz Analytics Machine Learning Graph Analytics Deep Learning Visualization GPU Memory 2 Data Processing Evolution Faster data access, less data movement Hadoop Processing, Reading from disk HDFS HDFS HDFS HDFS HDFS Read Query Write Read ETL Write Read ML Train Spark In-Memory Processing 25-100x Improvement Less code HDFS Language flexible Read Query ETL ML Train Primarily In-Memory Traditional GPU Processing 5-10x Improvement More code HDFS GPU CPU GPU CPU GPU ML Language rigid Query ETL Read Read Write Read Write Read Train Substantially on GPU 3 Data Movement and Transformation The bane of productivity and performance APP B Read Data APP B GPU APP B Copy & Convert Data CPU GPU Copy & Convert Copy & Convert APP A GPU Data APP A Load Data APP A 4 Data Movement and Transformation What if we could keep data on the GPU? APP B Read Data APP B GPU APP B Copy & Convert Data CPU GPU Copy & Convert Copy & Convert APP A GPU Data APP A Load Data APP A 5 Learning from Apache Arrow ● Each system has its own internal memory format ● All systems utilize the same memory format ● 70-80% computation wasted on serialization and deserialization ● No overhead for cross-system communication ● Similar functionality implemented in multiple projects ● Projects can share functionality (eg, Parquet-to-Arrow reader) From Apache Arrow -
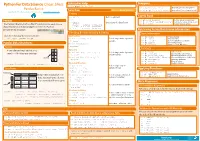
Cheat Sheet Pandas Python.Indd
Python For Data Science Cheat Sheet Asking For Help Dropping >>> help(pd.Series.loc) Pandas Basics >>> s.drop(['a', 'c']) Drop values from rows (axis=0) Selection Also see NumPy Arrays >>> df.drop('Country', axis=1) Drop values from columns(axis=1) Learn Python for Data Science Interactively at www.DataCamp.com Getting Sort & Rank >>> s['b'] Get one element -5 Pandas >>> df.sort_index() Sort by labels along an axis Get subset of a DataFrame >>> df.sort_values(by='Country') Sort by the values along an axis The Pandas library is built on NumPy and provides easy-to-use >>> df[1:] Assign ranks to entries Country Capital Population >>> df.rank() data structures and data analysis tools for the Python 1 India New Delhi 1303171035 programming language. 2 Brazil Brasília 207847528 Retrieving Series/DataFrame Information Selecting, Boolean Indexing & Setting Basic Information Use the following import convention: By Position (rows,columns) Select single value by row & >>> df.shape >>> import pandas as pd >>> df.iloc[[0],[0]] >>> df.index Describe index Describe DataFrame columns 'Belgium' column >>> df.columns Pandas Data Structures >>> df.info() Info on DataFrame >>> df.iat([0],[0]) Number of non-NA values >>> df.count() Series 'Belgium' Summary A one-dimensional labeled array a 3 By Label Select single value by row & >>> df.sum() Sum of values capable of holding any data type b -5 >>> df.loc[[0], ['Country']] Cummulative sum of values 'Belgium' column labels >>> df.cumsum() >>> df.min()/df.max() Minimum/maximum values c 7 Minimum/Maximum index -

Portable Stateful Big Data Processing in Apache Beam
Portable stateful big data processing in Apache Beam Kenneth Knowles Apache Beam PMC Software Engineer @ Google https://s.apache.org/ffsf-2017-beam-state [email protected] / @KennKnowles Flink Forward San Francisco 2017 Agenda 1. What is Apache Beam? 2. State 3. Timers 4. Example & Little Demo What is Apache Beam? TL;DR (Flink draws it more like this) 4 DAGs, DAGs, DAGs Apache Beam Apache Flink Apache Cloud Hadoop Apache Apache Dataflow Spark Samza MapReduce Apache Apache Apache (paper) Storm Gearpump Apex (incubating) FlumeJava (paper) Heron MillWheel (paper) Dataflow Model (paper) 2004 2005 2006 2007 2008 2009 2010 2011 2012 2013 2014 2015 2016 Apache Flink local, on-prem, The Beam Vision cloud Cloud Dataflow: Java fully managed input.apply( Apache Spark Sum.integersPerKey()) local, on-prem, cloud Sum Per Key Apache Apex Python local, on-prem, cloud input | Sum.PerKey() Apache Gearpump (incubating) ⋮ ⋮ 6 Apache Flink local, on-prem, The Beam Vision cloud Cloud Dataflow: Python fully managed input | KakaIO.read() Apache Spark local, on-prem, cloud KafkaIO Apache Apex ⋮ local, on-prem, cloud Apache Java Gearpump (incubating) class KafkaIO extends UnboundedSource { … } ⋮ 7 The Beam Model PTransform Pipeline PCollection (bounded or unbounded) 8 The Beam Model What are you computing? (read, map, reduce) Where in event time? (event time windowing) When in processing time are results produced? (triggers) How do refinements relate? (accumulation mode) 9 What are you computing? Read ParDo Grouping Composite Parallel connectors to Per element Group -

Pandas: Powerful Python Data Analysis Toolkit Release 0.25.3
pandas: powerful Python data analysis toolkit Release 0.25.3 Wes McKinney& PyData Development Team Nov 02, 2019 CONTENTS i ii pandas: powerful Python data analysis toolkit, Release 0.25.3 Date: Nov 02, 2019 Version: 0.25.3 Download documentation: PDF Version | Zipped HTML Useful links: Binary Installers | Source Repository | Issues & Ideas | Q&A Support | Mailing List pandas is an open source, BSD-licensed library providing high-performance, easy-to-use data structures and data analysis tools for the Python programming language. See the overview for more detail about whats in the library. CONTENTS 1 pandas: powerful Python data analysis toolkit, Release 0.25.3 2 CONTENTS CHAPTER ONE WHATS NEW IN 0.25.2 (OCTOBER 15, 2019) These are the changes in pandas 0.25.2. See release for a full changelog including other versions of pandas. Note: Pandas 0.25.2 adds compatibility for Python 3.8 (GH28147). 1.1 Bug fixes 1.1.1 Indexing • Fix regression in DataFrame.reindex() not following the limit argument (GH28631). • Fix regression in RangeIndex.get_indexer() for decreasing RangeIndex where target values may be improperly identified as missing/present (GH28678) 1.1.2 I/O • Fix regression in notebook display where <th> tags were missing for DataFrame.index values (GH28204). • Regression in to_csv() where writing a Series or DataFrame indexed by an IntervalIndex would incorrectly raise a TypeError (GH28210) • Fix to_csv() with ExtensionArray with list-like values (GH28840). 1.1.3 Groupby/resample/rolling • Bug incorrectly raising an IndexError when passing a list of quantiles to pandas.core.groupby. DataFrameGroupBy.quantile() (GH28113). -

Apache Flink Fast and Reliable Large-Scale Data Processing
Apache Flink Fast and Reliable Large-Scale Data Processing Fabian Hueske @fhueske 1 What is Apache Flink? Distributed Data Flow Processing System • Focused on large-scale data analytics • Real-time stream and batch processing • Easy and powerful APIs (Java / Scala) • Robust execution backend 2 What is Flink good at? It‘s a general-purpose data analytics system • Real-time stream processing with flexible windows • Complex and heavy ETL jobs • Analyzing huge graphs • Machine-learning on large data sets • ... 3 Flink in the Hadoop Ecosystem Libraries Dataflow Table API Table ML Library ML Gelly Library Gelly Apache MRQL Apache SAMOA DataSet API (Java/Scala) DataStream API (Java/Scala) Flink Core Optimizer Stream Builder Runtime Environments Embedded Local Cluster Yarn Apache Tez Data HDFS Hadoop IO Apache HBase Apache Kafka Apache Flume Sources HCatalog JDBC S3 RabbitMQ ... 4 Flink in the ASF • Flink entered the ASF about one year ago – 04/2014: Incubation – 12/2014: Graduation • Strongly growing community 120 100 80 60 40 20 0 Nov.10 Apr.12 Aug.13 Dec.14 #unique git committers (w/o manual de-dup) 5 Where is Flink moving? A "use-case complete" framework to unify batch & stream processing Data Streams • Kafka Analytical Workloads • RabbitMQ • ETL • ... • Relational processing Flink • Graph analysis • Machine learning “Historic” data • Streaming data analysis • HDFS • JDBC • ... Goal: Treat batch as finite stream 6 Programming Model & APIs HOW TO USE FLINK? 7 Unified Java & Scala APIs • Fluent and mirrored APIs in Java and Scala • Table API -

Arrow: Integration to 'Apache' 'Arrow'
Package ‘arrow’ September 5, 2021 Title Integration to 'Apache' 'Arrow' Version 5.0.0.2 Description 'Apache' 'Arrow' <https://arrow.apache.org/> is a cross-language development platform for in-memory data. It specifies a standardized language-independent columnar memory format for flat and hierarchical data, organized for efficient analytic operations on modern hardware. This package provides an interface to the 'Arrow C++' library. Depends R (>= 3.3) License Apache License (>= 2.0) URL https://github.com/apache/arrow/, https://arrow.apache.org/docs/r/ BugReports https://issues.apache.org/jira/projects/ARROW/issues Encoding UTF-8 Language en-US SystemRequirements C++11; for AWS S3 support on Linux, libcurl and openssl (optional) Biarch true Imports assertthat, bit64 (>= 0.9-7), methods, purrr, R6, rlang, stats, tidyselect, utils, vctrs RoxygenNote 7.1.1.9001 VignetteBuilder knitr Suggests decor, distro, dplyr, hms, knitr, lubridate, pkgload, reticulate, rmarkdown, stringi, stringr, testthat, tibble, withr Collate 'arrowExports.R' 'enums.R' 'arrow-package.R' 'type.R' 'array-data.R' 'arrow-datum.R' 'array.R' 'arrow-tabular.R' 'buffer.R' 'chunked-array.R' 'io.R' 'compression.R' 'scalar.R' 'compute.R' 'config.R' 'csv.R' 'dataset.R' 'dataset-factory.R' 'dataset-format.R' 'dataset-partition.R' 'dataset-scan.R' 'dataset-write.R' 'deprecated.R' 'dictionary.R' 'dplyr-arrange.R' 'dplyr-collect.R' 'dplyr-eval.R' 'dplyr-filter.R' 'expression.R' 'dplyr-functions.R' 1 2 R topics documented: 'dplyr-group-by.R' 'dplyr-mutate.R' 'dplyr-select.R' 'dplyr-summarize.R'