Roles of Specific Aminoglycoside-Ribosome Interactions in the Inhibition of Translation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Activity of Vancomycin Combined with Linezolid Against Clinical Vancomycin-Resistant Enterococcus Strains
Basic research Activity of vancomycin combined with linezolid against clinical vancomycin-resistant Enterococcus strains Gulseren Aktas Department of Medical Microbiology, Faculty of Medicine, Istanbul University, Corresponding author: Istanbul, Turkey Gulseren Aktas Department of Medical Submitted: 16 April 2019 Microbiology Accepted: 12 July 2019 Faculty of Medicine Istanbul University Arch Med Sci Istanbul, Turkey DOI: https://doi.org/10.5114/aoms.2020.96400 Phone: 090 212 Copyright © 2020 Termedia & Banach 4142000/32417 Fax: 090 212 4142037 E-mail: Abstract [email protected] Introduction: Because multi-drug-resistant Gram-positive bacteria have been isolated frequently worldwide and are difficult to treat, alternative treatment choices are required. Combination antibiotherapies have a distinct advan- tage over monotherapies in terms of their broad spectrum and synergistic effect. In the present study, it was aimed to investigate the in vitro activity of vancomycin combined with linezolid against clinical vancomycin-resistant enterococci (VRE) strains with high-level aminoglycoside resistance. Material and methods: A total of 30 randomly selected clinical VRE strains were studied. Susceptibility to agents tested was investigated using broth microdilution assay. The inoculum of strain was adjusted to approximately 5 × 105 CFU/ml in the wells. The results were interpreted in accordance with Clinical and Laboratory Standards Institute guidelines. In vitro activities of anti biotics in combination were assessed using the broth microcheckerboard technique. The fractional inhibitory concentration indexes (FICIs) were inter- preted as follows: synergism, FICI ≤ 0.5; additive/indifference, FICI ≤ 0.5 – ≤ 4; antagonism, FICI > 4. Results: All strains were resistant to vancomycin and susceptible to linezolid. The MIC50,90 and MICrange values of antimicrobials were 512, 512, and 512–1024 µg/ ml for vancomycin; 2, 2, and 2–4 µg/ml for linezolid. -

Farrukh Javaid Malik
I Farrukh Javaid Malik THESIS PRESENTED TO OBTAIN THE GRADE OF DOCTOR OF THE UNIVERSITY OF BORDEAUX Doctoral School, SP2: Society, Politic, Public Health Specialization Pharmacoepidemiology and Pharmacovigilance By Farrukh Javaid Malik “Analysis of the medicines panorama in Pakistan – The case of antimicrobials: market offer width and consumption.” Under the direction of Prof. Dr. Albert FIGUERAS Defense Date: 28th November 2019 Members of Jury M. Francesco SALVO, Maître de conférences des universités – praticien hospitalier, President Université de Bordeaux M. Albert FIGUERAS, Professeur des universités – praticien hospitalier, Director Université Autonome de Barcelone Mme Antonia AGUSTI, Professeure, Vall dʹHebron University Hospital Referee Mme Montserrat BOSCH, Praticienne hospitalière, Vall dʹHebron University Hospital Referee II Abstract A country’s medicines market is an indicator of its healthcare system, the epidemiological profile, and the prevalent practices therein. It is not only the first logical step to study the characteristics of medicines authorized for marketing, but also a requisite to set up a pharmacovigilance system, thus promoting rational drug utilization. The three medicines market studies presented in the present document were conducted in Pakistan with the aim of describing the characteristics of the pharmaceutical products available in the country as well as their consumption at a national level, with a special focus on antimicrobials. The most important cause of antimicrobial resistance is the inappropriate consumption of antimicrobials. The results of the researches conducted in Pakistan showed some market deficiencies which could be addressed as part of the national antimicrobial stewardship programmes. III Résumé Le marché du médicament d’un pays est un indicateur de son système de santé, de son profil épidémiologique et des pratiques [de prescription] qui y règnent. -

Antibiotic Use in South Korea from 2007 to 2014: a Health Insurance Database-Generated Time Series Analysis
RESEARCH ARTICLE Antibiotic use in South Korea from 2007 to 2014: A health insurance database-generated time series analysis Juhee Park1☯, Euna Han2☯, Soo Ok Lee1, Dong-Sook Kim1* 1 Department of Research, Health Insurance Review & Assessment Service, Wonju, Korea, 2 College of Pharmacy, Yonsei Institute of Pharmaceutical Science, Yonsei University, Seoul, Korea ☯ These authors contributed equally to this work. * [email protected] Abstract a1111111111 a1111111111 a1111111111 Background a1111111111 a1111111111 Inappropriate antibiotic use significantly contributes to antibiotic-resistance, resulting in reduced antibiotic efficacy and increasing physical burden and cost of disease. The goal of this study was to explore antibiotic usage patterns in South Korea using 2007±2014 health insurance claims data. OPEN ACCESS Methods Citation: Park J, Han E, Lee SO, Kim D-S (2017) Antibiotic use in South Korea from 2007 to 2014: A We used the Health Insurance Review & Assessment Service data, which represents nearly health insurance database-generated time series the entire population of South Korea, to discern patterns in antibiotic prescribing practices. analysis. PLoS ONE 12(5): e0177435. https://doi. The daily dose, as defined by the World Health Organization ([defined daily doses]/1000 org/10.1371/journal.pone.0177435 inhabitants/day, [DID]), was used as a measure of antibiotic use. Subgroup analyses were Editor: Yeng-Tseng Wang, Kaohsiung Medical performed on the basis of patient characteristics (sex, age, and disease) and provider char- University, TAIWAN acteristics (type of medical institution). Received: July 21, 2016 Accepted: April 20, 2017 Results Published: May 17, 2017 Antibiotic use in DID increased from 23.5 in 2007 to 27.7 in 2014. -

Treatment with Sub-Inhibitory Kanamycin Induces Adaptive Resistance to Aminoglycoside Antibiotics Via the Acrd Multidrug Efflux Pump in Escherichia Coli K-12
Journal of Experimental Microbiology and Immunology (JEMI) Vol. 16: 11 – 16 Copyright © April 2012, M&I UBC Treatment with Sub-inhibitory Kanamycin Induces Adaptive Resistance to Aminoglycoside Antibiotics via the AcrD Multidrug Efflux Pump in Escherichia coli K-12 Kiran Sidhu, Martha Talbot, Kirstin Van Mil, and Meghan Verstraete Department of Microbiology & Immunology, University of British Columbia Research has shown that exposing Escherichia coli cells to sub-inhibitory concentrations of kanamycin induces adaptive resistance upon subsequent exposure to lethal levels of both structurally-similar and unrelated antibiotics. AcrD is an efflux pump that forms a complex with the outer membrane pore TolC and the periplasmic membrane fusion protein AcrA. Together, this complex works to export a variety of aminoglycosides and amphiphilic compounds from the cell. The involvement of AcrD in the observed adaptive resistance was assessed by comparing the induction of adaptive resistance upon exposure to kanamycin in wild- type and acrD-deficient Escherichia coli strains. Both strains were pretreated with sub- inhibitory concentrations of kanamycin and subsequently exposed to lethal concentrations of ampicillin, kanamycin, nalidixic acid, streptomycin, and tetracycline. Growth was monitored over 18 hours via optical density readings. Comparing the relative growth of the different treatments revealed that adaptive resistance was only induced in the wild-type strain towards kanamycin and streptomycin. This indicates that AcrD is necessary for the induced adaptive resistance and that it is specific to aminoglycosides. Additionally, molecular techniques were used to assess the level of expression of acrA, which was found to be upregulated in both the wild-type and acrD deletion strains following pretreatment with kanamycin. -

TETRACYCLINES and CHLORAMPHENICOL Protein Synthesis
ANTIMICROBIALS INHIBITING PROTEIN SYNTHESIS AMINOGLYCOSIDES MACROLIDES TETRACYCLINES AND CHLORAMPHENICOL Protein synthesis Aminoglycosides 1. Aminoglycosides are group of natural and semi -synthetic antibiotics. They have polybasic amino groups linked glycosidically to two or more aminosugar like: sterptidine, 2-deoxy streptamine, glucosamine 2. Aminoglycosides which are derived from: Streptomyces genus are named with the suffix –mycin. While those which are derived from Micromonospora are named with the suffix –micin. Classification of Aminoglycosides 1. Systemic aminogycosides Streptomycin (Streptomyces griseus) Gentamicin (Micromonospora purpurea) Kanamycin (S. kanamyceticus) Tobramycin (S. tenebrarius) Amikacin (Semisynthetic derivative of Kanamycin) Sisomicin (Micromonospora inyoensis) Netilmicin (Semisynthetic derivative of Sisomicin) 2. Topical aminoglycosides Neomycin (S. fradiae) Framycetin (S. lavendulae) Pharmacology of Streptomycin NH H2N NH HO OH Streptidine OH NH H2N O O NH CHO L-Streptose CH3 OH O HO O HO NHCH3 N-Methyl-L- Glucosamine OH Streptomycin Biological Source It is a oldest aminoglycoside antibiotic obtained from Streptomyces griseus. Antibacterial spectrum 1. It is mostly active against gram negative bacteria like H. ducreyi, Brucella, Yersinia pestis, Francisella tularensis, Nocardia,etc. 2. It is also used against M.tuberculosis 3. Few strains of E.coli, V. cholerae, H. influenzae , Enterococci etc. are sensitive at higher concentration. Mechanism of action Aminoglycosides bind to the 16S rRNA of the 30S subunit and inhibit protein synthesis. 1. Transport of aminoglycoside through cell wall and cytoplasmic membrane. a) Diffuse across cell wall of gram negative bacteria by porin channels. b) Transport across cell membrane by carrier mediated process liked with electron transport chain 2. Binding to ribosome resulting in inhibition of protein synthesis A. -

Pharmacokinetic Training Packet for Pharmacists
Pharmacokinetic Training Packet for Pharmacists Revised 1/09, 6/12 Original document compiled by: Elizabeth D. Hermsen, Pharm.D., M.B.A., BCPS-ID Updated by: Alan Gross, Pharm.D., BCPS Thanks to Erin Iselin, Scott McMullen, Chris Shaffer, & Keith Olsen for your thoughtful review! Any questions? Call or email Alan Gross at 559-4149/[email protected] Table of Contents Pharmacokinetic definitions and principles 3 Aminoglycoside overview 4 Extended-interval (Once daily) aminoglycoside dosing 8 Aminoglycoside pharmacokinetic calculations 10 Aminoglycoside dosing in patients with cystic fibrosis 12 Vancomycin overview and pharmacokinetic calculations 15 Clinical Pearls 21 Dialysis – Aminoglycosides and Vancomycin 21 TNMC Nephrology Protocol for Vancomycin Dosing 21 Clinical Pharmacokinetic Consult Service 23 2 Pharmacokinetic Definitions and Principles Kel, Ke, or Kd or Elimination Rate Constant 1 • The fraction or percentage of the total amount of drug in the body eliminated per unit of time. • Estimated with 2 drug levels taken between doses (the slope of the line). To be accurate, 2-4 half-lives should occur between the levels.1 -kel(τ) • In pharmacokinetic calculations, the term e represents the fraction of the serum concentration that remains. Thus, 1 - e- kel(τ) represents the fraction of the serum concentration that is eliminated. t 1/2 or Half-life 1 • The time required for the TOTAL amount of remaining drug in the body to decline by 50%. • Sometimes referred to as β t ½ to distinguish it from the distribution half-life, α t ½, used in two compartment modeling.1 Peak, C max1 • C max is the maximum measurable drug concentration at the end of an infusion BEFORE significant distribution occurs. -

Aminoglycoside Allergic Reactions
pharmacy Review Aminoglycoside Allergic Reactions Lindsey M. Childs-Kean 1,* , Kristy M. Shaeer 2, Sheeba Varghese Gupta 3 and Jonathan C. Cho 4 1 Department of Pharmacotherapy and Translational Research, University of Florida College of Pharmacy, Gainesville, FL 32610, USA 2 Department of Pharmacotherapeutics and Clinical Research, University of South Florida College of Pharmacy, Tampa, FL 33612, USA 3 Department of Pharmaceutical Sciences, University of South Florida College of Pharmacy, Tampa, FL 33612, USA 4 Department of Clinical Sciences, Ben and Maytee Fisch College of Pharmacy, The University of Texas at Tyler, Tyler, TX 75799, USA * Correspondence: [email protected]fl.edu Received: 13 July 2019; Accepted: 25 August 2019; Published: 29 August 2019 Abstract: Aminoglycosides are antimicrobial agents that are primarily used for infections caused by Gram-negative pathogens. The purpose of this article is to review the allergic reactions reported in the published literature to aminoglycoside antibiotics. A thorough PubMed search was conducted and excluded non-allergic adverse reactions to aminoglycosides. Allergic reactions to aminoglycosides occur infrequently, but can include cutaneous reactions as well as systemic reactions, including anaphylaxis. Of the evaluated aminoglycosides, gentamicin had the most reported allergic reactions, including the most reports of anaphylaxis, followed by tobramycin, and then amikacin. Most reports of allergic reactions occurred in patients who had a prior exposure to some dosage form of an aminoglycoside. Cross-reactivity among aminoglycosides is common and occurs due to the similarities in their chemical structures. Desensitization protocols to tobramycin have been described in the literature. Keywords: aminoglycosides; allergy; hypersensitivity; gentamicin; tobramycin; amikacin 1. Introduction Antibiotics are one of the most common causes of life-threatening medication allergies [1]. -

Aminoglycoside Dosing and Monitoring Guidelines for Adult Patients at Stony Brook University Hospital
Aminoglycoside Dosing and Monitoring Guidelines for Adult Patients at Stony Brook University Hospital Algorithm for initial Aminoglycoside dosing Type of Infection Gram-negative Gram- Infection positive Infection Stable Renal Unstable Renal HD or Dosing for Function Function CVVHD Synergy (see pg.13- Cr Cl or eGFR * Conventional 14) Cr Cl or eGFR * Conventional ≥ 40 ml/min Dosing Dosing < 40 ml/min (see pg. 8-10) (see pg 11-12) Extended Interval Conventional (see pg. 4-7) Dosing (also known as (see pg. 8-10) Once-Daily Dosing) * eGFR is normalized to BSA 1.73m2. Convert normalized eGFR (ml/min/1.73m2) to individualized eGFR (ml/min) by multiplying eGFR (ml/min/1.73m2) with Patient’s BSA and divided by 1.73. Both estimated Cr Cl by Cockcroft-Gault equation and eGFR by CKD-EPI equation provide reasonable estimates for aminoglycoside dosing. However, due to the limitations of these estimating equations, it would be prudent for clinicians to apply clinical judgment when interpreting either estimates (Cr Cl or eGFR) in elderly patients, patients at the extremes of muscle mass and patients with unstable serum creatinine levels. If the 2 estimates (Cr Cl and eGFR) lead to different dosing regimens, it would be advisable for clinicians to incorporate risk versus benefit assessment in determining the aminoglycoside dosing regimen. Author: Melinda Monteforte, B.S., PharmD., BCPS, AQ-ID Date: 01/2015 Reviewed by Pharmacy Department, Infectious Diseases Division, Nephrology Division, Antimicrobial Stewardship Committee Page 1 of 14 Disclaimer The information contained is adapted from published pharmacokinetic and pharmacodynamics literature The information provided is not intended to replace sound clinical judgment This is not intended to be used for follow up when measured serum concentrations are available. -
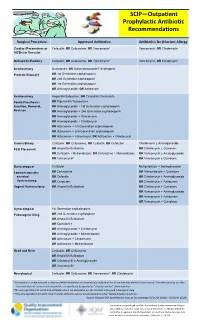
SCIP—Outpatient Prophylactic Antibiotic Recommendations
SCIP—Outpatient Prophylactic Antibiotic Recommendations Surgical Procedure Approved Antibiotics Antibiotics for β-lactam Allergy Cardiac (Pacemakers or Cefazolin; OR Cefuroxime; OR Vancomycin* Vancomycin; OR Clindamycin AICDs) or Vascular Orthopedic/Podiatry Cefazolin; OR Cefuroxime; OR Vancomycin* Vancomycin; OR Clindamycin Genitourinary Quinolone†; OR Sulfamethoxazole/Trimethoprim Prostate Biopsy†† OR 1st Generation cephalosporin OR 2nd Generation cephaloporin OR 3rd Generation cephalosporin OR Aminoglycoside; OR Aztreonam Genitourinary Ampicillin/Sulbactam; OR Ticarcillin/Clavulanate Penile Prosthesis - OR Piperacillin/Tazobactam Insertion, Removal, OR Aminoglycoside + 1st Generation cephalosporin Revision OR Aminoglycoside + 2nd Generation cephalosporin OR Aminoglycoside + Vancomycin OR Aminoglycoside + Clindamycin OR Aztreonam + 1st Generation cephalosporin OR Aztreonam + 2nd Generation cephalosporin OR Aztreonam + Vancomycin; OR Aztreonam + Clindamycin Gastric/Biliary Cefazolin; OR Cefuroxime; OR Cefoxitin; OR Cefotetan Clindamycin ± Aminoglycoside PEG Placement OR Ampicillin/Sulbactam OR Clindamycin ± Quinolone OR Cefazolin + Metronidazole; OR Cefurozime + Metronidazole OR Vancomycin ± Aminoglycoside OR Vancomycin* OR Vancomycin ± Quinolone Gynecological Cefazolin Metronidazole + Aminoglycoside Laparoscopically- OR Cefuroxime OR Metronidazole + Quinolone assisted OR Cefoxitin OR Clindamycin + Aminoglycoside Hysterectomy, OR Cefotetan OR Clindamycin + Aztreonam Vaginal Hysterectomy OR Ampicillin/Sulbactam OR Clindamycin + Quinolone -

Retrospective Study of Outcome in Patients Treated for Staphylococcus
ORIGINAL ARTICLE Retrospective study of outcome in patients treated for Staphy 1ococcus aureus b ac teremia Pauline E. Gosdenl, Barnaby C. Reeves2,]ames R. S. Osborne3, Andrew Turner' and Michael R. Millarl 'Department of mcrobiology, 2Research and Development Support Unit, and 3Department of Pathology, University of Bristol, Bristol Royal Infirmary, United Bristol Healthcare Trust, Bristol, UK Objective: To investigate whether a change in current treatment practice for Staphylococcus aureus bacteremia from flucloxacillin and aminoglycoside to flucloxacillin and fusidic acid was associated with any changes in outcome. Methods: A retrospective analysis was carried out of 316 episodes of S. aureus bacteremia diagnosed and treated in a tertiary hospital complex between 1983 and 1993. Outcomes considered were (1) death related to the infection and (2) relapse following cessation of antibiotic therapy. Results: Mortality related to infection, which occurred in 24% of patients, was unrelated to treatment with the combination of flucloxacillin and fusidic acid; however, increasing age was a significant risk factor (OR per decade = 1.35, 95% CI = 1.1&1.55), and increasing duration of treatment (OR per week of treatment = 0.63,95% CI = 0.52-0.77), use of flucloxacillin (OR= 0.30,95% CI = 0.14-0.64). presence of an intravascular device (OR= 0.39,95% CI = 0.20-0.78) and presence of a skin lesion (OR = 0.51, 95% CI = 0.26-0.99) were significant protective factors. The only factor significantly related to relapse, which occurred in 11% of patients, was treatment with the combination of flucloxacillin and fusidic acid (OR = 0.32, 95% CI = 0.12-0.85). -

2. Fusidic Acid in Skin and Soft-Tissue Infections
Acta Derm Venereol 2008; Suppl 216: 14–20 2. Fusidic acid in skin and soft-tissue infections Barry H. LONG Topical antibacterial therapy is an important component treatment may subsequently require modification once in managing skin and soft-tissue infections (SSTIs). the culture results become available. Fusidic acid, a narrow-spectrum antibiotic active against Topical antibacterials have a distinct advantage over Staphylococcus aureus, has shown good skin permeabi- systemic agents, in that they can be applied to the af- lity and low allergenic potential. The resistance rate in fected area and therefore high local concentrations of the S. aureus remains low, as shown in a study of Canadian agent may be achieved. With selection of the appropriate hospitals from 1999 to 2005. In treating primary skin agent, interaction with normal flora can be avoided. The infections, including impetigo, fusidic acid cream and ideal topical antibiotic should: ointment provided similar response rates and equal/better • have a selective effect on one (or at least very few) tolerability compared with other topical and oral anti- organisms of the same class, therefore minimizing biotics. Fusidic acid and mupirocin are equally or more the development of cross-resistance to other orga- efficacious than oral treatment in localized impetigo, nisms; and may be similarly efficacious in extensive impetigo, • not cause allergic reactions or potential cross-allergic according to a recent Cochrane review. In clinical prac- reactions with other medications of the same class or tice, mupirocin is often reserved for methicillin-resistant individual components of these, such as preservati- S. aureus infections. Studies of oral fusidic acid forms in ves; SSTI have shown that: tablets are as effective as compara- • be safe, efficacious and ideally penetrate the skin in tor antibiotics; they have fewer side-effects; a suspension sufficiently high concentrations to kill bacteria effic- achieves high cure rates, and is suitable for paediatric iently; use. -
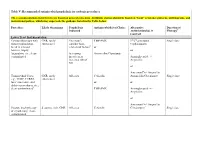
Table V: Recommended Antimicrobial Prophylaxis for Urologic Procedures
Table V: Recommended antimicrobial prophylaxis for urologic procedures The recommendations listed herein are based on general consensus. Antibiotic choices should be based on “local” resistance patterns, antibiograms, and institutional polices, which may supersede the guidance listed in the Table below. Procedure Likely Organisms Prophylaxis Antimicrobial(s) of Choice Alternative Duration of Indicated Antimicrobial(s), if Therapy¶ required Lower Tract Instrumentation Cystourethroscopy with GNR, rarely Uncertain§; TMP-SMX 1st/2nd generation Single dose minor manipulation, enterococci† consider host- Cephalosporin break in mucosal related risk factors‡ or barriers, biopsy, or fulguration, etc.; clean- Increasing Amoxicillin/Clavulanate contaminated invasiveness Aminoglycoside +/- increases risk of Ampicillin SSI or Aztreonam¥ +/- Ampicillin Transurethral Cases: GNR, rarely All cases Cefazolin Amoxicillin/Clavulanate Single dose e.g., TURP, TURBT, enterococci laser enucleative and or or ablative procedures, etc.; clean-contaminatedǁ TMP-SMX Aminoglycoside +/- Ampicillin or Aztreonam¥ +/- Ampicillin Prostate brachytherapy S. aureus, skin; GNR All cases Cefazolin Clindamycin** Single dose or cryotherapy; clean- contaminated Lower Tract Instrumentation Transrectal prostate GNR, anaerobes††; All cases Fluoroquinolone Aztreonam Single dose biopsy; contaminated consider multi-drug resistance coverage, or if risks of systemic May need to consider antibiotics within six 1st/2nd gen. Cephalosporin infectious disease months, international +/- Aminoglycoside