Nonparametric Spatial Covariance Functions: Estimation and Testing
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Nonparametric Regression 1 Introduction
Nonparametric Regression 10/36-702 Larry Wasserman 1 Introduction Now we focus on the following problem: Given a sample (X1;Y1);:::,(Xn;Yn), where d Xi 2 R and Yi 2 R, estimate the regression function m(x) = E(Y jX = x) (1) without making parametric assumptions (such as linearity) about the regression function m(x). Estimating m is called nonparametric regression or smoothing. We can write Y = m(X) + where E() = 0. This follows since, = Y − m(X) and E() = E(E(jX)) = E(m(X) − m(X)) = 0 A related problem is nonparametric prediction. Given a pair (X; Y ), we want to predict Y from X. The optimal predictor (under squared error loss) is the regression function m(X). Hence, estimating m is of interest for its own sake and for the purposes of prediction. Example 1 Figure 1 shows data on bone mineral density. The plots show the relative change in bone density over two consecutive visits, for men and women. The smooth estimates of the regression functions suggest that a growth spurt occurs two years earlier for females. In this example, Y is change in bone mineral density and X is age. Example 2 Figure 2 shows an analysis of some diabetes data from Efron, Hastie, Johnstone and Tibshirani (2004). The outcome Y is a measure of disease progression after one year. We consider four covariates (ignoring for now, six other variables): age, bmi (body mass index), and two variables representing blood serum measurements. A nonparametric regression model in this case takes the form Y = m(x1; x2; x3; x4) + . -

Nonparametric Regression with Complex Survey Data
CHAPTER 11 NONPARAMETRIC REGRESSION WITH COMPLEX SURVEY DATA R. L. Chambers Department of Social Statistics University of Southampton A.H. Dorfman Office of Survey Methods Research Bureau of Labor Statistics M.Yu. Sverchkov Department of Statistics Hebrew University of Jerusalem June 2002 11.1 Introduction The problem considered here is one familiar to analysts carrying out exploratory data analysis (EDA) of data obtained via a complex sample survey design. How does one adjust for the effects, if any, induced by the method of sampling used in the survey when applying EDA methods to these data? In particular, are adjustments to standard EDA methods necessary when the analyst's objective is identification of "interesting" population (rather than sample) structures? A variety of methods for adjusting for complex sample design when carrying out parametric inference have been suggested. See, for example, SHS, Pfeffermann (1993) and Breckling et al (1994). However, comparatively little work has been done to date on extending these ideas to EDA, where a parametric formulation of the problem is typically inappropriate. We focus on a popular EDA technique, nonparametric regression or scatterplot smoothing. The literature contains a limited number of applications of this type of analysis to survey data, usually based on some form of sample weighting. The design-based theory set out in chapter 10, with associated references, provides an introduction to this work. See also Chesher (1997). The approach taken here is somewhat different. In particular, it is model-based, building on the sample distribution concept discussed in section 2.3. Here we develop this idea further, using it to motivate a number of methods for adjusting for the effect of a complex sample design when estimating a population regression function. -

Nonparametric Bayesian Inference
Nonparametric Bayesian Methods 1 What is Nonparametric Bayes? In parametric Bayesian inference we have a model M = ff(yjθ): θ 2 Θg and data Y1;:::;Yn ∼ f(yjθ). We put a prior distribution π(θ) on the parameter θ and compute the posterior distribution using Bayes' rule: L (θ)π(θ) π(θjY ) = n (1) m(Y ) Q where Y = (Y1;:::;Yn), Ln(θ) = i f(Yijθ) is the likelihood function and n Z Z Y m(y) = m(y1; : : : ; yn) = f(y1; : : : ; ynjθ)π(θ)dθ = f(yijθ)π(θ)dθ i=1 is the marginal distribution for the data induced by the prior and the model. We call m the induced marginal. The model may be summarized as: θ ∼ π Y1;:::;Ynjθ ∼ f(yjθ): We use the posterior to compute a point estimator such as the posterior mean of θ. We can also summarize the posterior by drawing a large sample θ1; : : : ; θN from the posterior π(θjY ) and the plotting the samples. In nonparametric Bayesian inference, we replace the finite dimensional model ff(yjθ): θ 2 Θg with an infinite dimensional model such as Z F = f : (f 00(y))2dy < 1 (2) Typically, neither the prior nor the posterior have a density function with respect to a dominating measure. But the posterior is still well defined. On the other hand, if there is a dominating measure for a set of densities F then the posterior can be found by Bayes theorem: R A Ln(f)dπ(f) πn(A) ≡ P(f 2 AjY ) = R (3) F Ln(f)dπ(f) Q where A ⊂ F, Ln(f) = i f(Yi) is the likelihood function and π is a prior on F. -
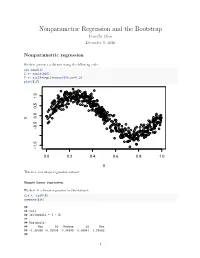
Nonparametric Regression and the Bootstrap Yen-Chi Chen December 5, 2016
Nonparametric Regression and the Bootstrap Yen-Chi Chen December 5, 2016 Nonparametric regression We first generate a dataset using the following code: set.seed(1) X <- runif(500) Y <- sin(X*2*pi)+rnorm(500,sd=0.2) plot(X,Y) 1.0 0.5 0.0 Y −0.5 −1.5 0.0 0.2 0.4 0.6 0.8 1.0 X This is a sine shape regression dataset. Simple linear regression We first fit a linear regression to this dataset: fit <- lm(Y~X) summary(fit) ## ## Call: ## lm(formula = Y ~ X) ## ## Residuals: ## Min 1Q Median 3Q Max ## -1.38395 -0.35908 0.04589 0.38541 1.05982 ## 1 ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 1.03681 0.04306 24.08 <2e-16 *** ## X -1.98962 0.07545 -26.37 <2e-16 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 '' 1 ## ## Residual standard error: 0.4775 on 498 degrees of freedom ## Multiple R-squared: 0.5827, Adjusted R-squared: 0.5819 ## F-statistic: 695.4 on 1 and 498 DF, p-value: < 2.2e-16 We observe a negative slope and here is the scatter plot with the regression line: plot(X,Y, pch=20, cex=0.7, col="black") abline(fit, lwd=3, col="red") 1.0 0.5 0.0 Y −0.5 −1.5 0.0 0.2 0.4 0.6 0.8 1.0 X After fitted a regression, we need to examine the residual plot to see if there is any patetrn left in the fit: plot(X, fit$residuals) 2 1.0 0.5 0.0 fit$residuals −0.5 −1.0 0.0 0.2 0.4 0.6 0.8 1.0 X We see a clear pattern in the residual plot–this suggests that there is a nonlinear relationship between X and Y. -

Nonparametric Regression
Nonparametric Regression Statistical Machine Learning, Spring 2015 Ryan Tibshirani (with Larry Wasserman) 1 Introduction, and k-nearest-neighbors 1.1 Basic setup, random inputs Given a random pair (X; Y ) Rd R, recall that the function • 2 × f0(x) = E(Y X = x) j is called the regression function (of Y on X). The basic goal in nonparametric regression is ^ d to construct an estimate f of f0, from i.i.d. samples (x1; y1);::: (xn; yn) R R that have the same joint distribution as (X; Y ). We often call X the input, predictor,2 feature,× etc., and Y the output, outcome, response, etc. Importantly, in nonparametric regression we do not assume a certain parametric form for f0 Note for i.i.d. samples (x ; y );::: (x ; y ), we can always write • 1 1 n n yi = f0(xi) + i; i = 1; : : : n; where 1; : : : n are i.i.d. random errors, with mean zero. Therefore we can think about the sampling distribution as follows: (x1; 1);::: (xn; n) are i.i.d. draws from some common joint distribution, where E(i) = 0, and then y1; : : : yn are generated from the above model In addition, we will assume that each i is independent of xi. As discussed before, this is • actually quite a strong assumption, and you should think about it skeptically. We make this assumption for the sake of simplicity, and it should be noted that a good portion of theory that we cover (or at least, similar theory) also holds without the assumption of independence between the errors and the inputs 1.2 Basic setup, fixed inputs Another common setup in nonparametric regression is to directly assume a model • yi = f0(xi) + i; i = 1; : : : n; where now x1; : : : xn are fixed inputs, and 1; : : : are still i.i.d. -

Testing for No Effect in Nonparametric Regression Via Spline Smoothing Techniques
Ann. Inst. Statist. Math. Vol. 46, No. 2, 251-265 (1994) TESTING FOR NO EFFECT IN NONPARAMETRIC REGRESSION VIA SPLINE SMOOTHING TECHNIQUES JUEI-CHAO CHEN Graduate Institute of Statistics and Actuarial Science, Feng Chic University, Taiehung ~072~, Taiwan, ROC (Received June 8, 1992; revised August 2, 1993) Abstract. We propose three statistics for testing that a predictor variable has no effect on the response variable in regression analysis. The test statistics are integrals of squared derivatives of various orders of a periodic smoothing spline fit to the data. The large sample properties of the test statistics are investigated under the null hypothesis and sequences of local alternatives and a Monte Carlo study is conducted to assess finite sample power properties. Key words and phrases: Asymptotic distribution, local alternatives, nonpara- metric regression, Monte Carlo. 1. Introduction Regression analysis is used to study relationships between the response vari- able and a predictor variable; therefore, all of the inference is based on the as- sumption that the response variable actually depends on the predictor variable. Testing for no effect is the same as checking this assumption. In this paper, we propose three statistics for testing that a predictor variable has no effect on the response variable and derive their asymptotic distributions. Suppose we have an experiment which yields observations (tin,y1),..., (tn~, Yn), where yj represents the value of a response variable y at equally spaced values tjn = (j - 1)/n, j = 1,...,n, of the predictor variable t. The regression model is (1.1) yj = #(tin) + ey, j --- 1,...,n, where the ej are iid random errors with E[el] = 0, Var[r ~2 and 0 K E[e 41 K oe. -

Chapter 5 Nonparametric Regression
Chapter 5 Nonparametric Regression 1 Contents 5 Nonparametric Regression 1 1 Introduction . 3 1.1 Example: Prestige data . 3 1.2 Smoothers . 4 2 Local polynomial regression: Lowess . 4 2.1 Window-span . 6 2.2 Inference . 7 3 Kernel smoothing . 9 4 Splines . 10 4.1 Number and position of knots . 11 4.2 Smoothing splines . 12 5 Penalized splines . 15 2 1 Introduction A linear model is desirable because it is simple to fit, it is easy to understand, and there are many techniques for testing the assumptions. However, in many cases, data are not linearly related, therefore, we should not use linear regression. The traditional nonlinear regression model fits the model y = f(Xβ) + 0 where β = (β1, . βp) is a vector of parameters to be estimated and X is the matrix of predictor variables. The function f(.), relating the average value of the response y on the predictors, is specified in advance, as it is in a linear regression model. But, in some situa- tions, the structure of the data is so complicated, that it is very difficult to find a functions that estimates the relationship correctly. A solution is: Nonparametric regression. The general nonparametric regression model is written in similar manner, but f is left unspecified: y = f(X) + = f(x1,... xp) + Most nonparametric regression methods assume that f(.) is a smooth, continuous function, 2 and that i ∼ NID(0, σ ). An important case of the general nonparametric model is the nonparametric simple regres- sion, where we only have one predictor y = f(x) + Nonparametric simple regression is often called scatterplot smoothing, because an important application is to tracing a smooth curve through a scatterplot of y against x and display the underlying structure of the data. -

Nonparametric-Regression.Pdf
Nonparametric Regression John Fox Department of Sociology McMaster University 1280 Main Street West Hamilton, Ontario Canada L8S 4M4 [email protected] February 2004 Abstract Nonparametric regression analysis traces the dependence of a response variable on one or several predictors without specifying in advance the function that relates the predictors to the response. This article dis- cusses several common methods of nonparametric regression, including kernel estimation, local polynomial regression, and smoothing splines. Additive regression models and semiparametric models are also briefly discussed. Keywords: kernel estimation; local polynomial regression; smoothing splines; additive regression; semi- parametric regression Nonparametric regression analysis traces the dependence of a response variable (y)ononeorseveral predictors (xs) without specifying in advance the function that relates the response to the predictors: E(yi)=f(x1i,...,xpi) where E(yi) is the mean of y for the ith of n observations. It is typically assumed that the conditional variance of y,Var(yi|x1i,...,xpi) is a constant, and that the conditional distribution of y is normal, although these assumptions can be relaxed. Nonparametric regression is therefore distinguished from linear regression, in which the function relating the mean of y to the xs is linear in the parameters, E(yi)=α + β1x1i + ···+ βpxpi and from traditional nonlinear regression, in which the function relating the mean of y to the xs, though nonlinear in its parameters, is specified explicitly, E(yi)=f(x1i,...,xpi; γ1,...,γk) In traditional regression analysis, the object is to estimate the parameters of the model – the βsorγs. In nonparametric regression, the object is to estimate the regression function directly. -

Lecture 8: Nonparametric Regression 8.1 Introduction
STAT/Q SCI 403: Introduction to Resampling Methods Spring 2017 Lecture 8: Nonparametric Regression Instructor: Yen-Chi Chen 8.1 Introduction Let (X1;Y1); ··· ; (Xn;Yn) be a bivariate random sample. In the regression analysis, we are often interested in the regression function m(x) = E(Y jX = x): Sometimes, we will write Yi = m(Xi) + i; where i is a mean 0 noise. The simple linear regression model is to assume that m(x) = β0 + β1x, where β0 and β1 are the intercept and slope parameter. In this lecture, we will talk about methods that direct estimate the regression function m(x) without imposing any parametric form of m(x). Given a point x0, assume that we are interested in the value m(x0). Here is a simple method to estimate that value. When m(x0) is smooth, an observation Xi ≈ x0 implies m(Xi) ≈ m(x0). Thus, the response value Yi = m(Xi) + i ≈ m(x0) + i. Using this observation, to reduce the noise i, we can use the sample average. Thus, an estimator of m(x0) is to take the average of those responses whose covariate are close to x0. To make it more concrete, let h > 0 be a threshold. The above procedure suggests to use P n Yi P i:jXi−x0|≤h i=1 YiI(jXi − x0j ≤ h) mb loc(x0) = = Pn ; (8.1) nh(x0) i=1 I(jXi − x0j ≤ h) where nh(x0) is the number of observations where the covariate X : jXi − x0j ≤ h. This estimator, mb loc, is called the local average estimator. -

Nonparametric Regression in R
Nonparametric Regression in R An Appendix to An R Companion to Applied Regression, third edition John Fox & Sanford Weisberg last revision: 2018-09-26 Abstract In traditional parametric regression models, the functional form of the model is specified before the model is fit to data, and the object is to estimate the parameters of the model. In nonparametric regression, in contrast, the object is to estimate the regression function directly without specifying its form explicitly. In this appendix to Fox and Weisberg (2019), we describe how to fit several kinds of nonparametric-regression models in R, including scatterplot smoothers, where there is a single predictor; models for multiple regression; additive regression models; and generalized nonparametric-regression models that are analogs to generalized linear models. 1 Nonparametric Regression Models The traditional nonlinear regression model that is described in the on-line appendix to the R Com- panion on nonlinear regression fits the model y = m(x; θ) + " where θ is a vector of parameters to be estimated, and x is a vector of predictors;1 the errors " are assumed to be normally and independently distributed with mean 0 and constant variance σ2. The function m(x; θ), relating the average value of the response y to the predictors, is specified in advance, as it is in a linear regression model. The general nonparametric regression model is written in a similar manner, but the function m is left unspecified: y = m(x) + " = m(x1; x2; : : : ; xp) + " 0 for the p predictors x = (x1; x2; : : : ; xp) . Moreover, the object of nonparametric regression is to estimate the regression function m(x) directly, rather than to estimate parameters. -

Parametric Versus Semi and Nonparametric Regression Models
Parametric versus Semi and Nonparametric Regression Models Hamdy F. F. Mahmoud1;2 1 Department of Statistics, Virginia Tech, Blacksburg VA, USA. 2 Department of Statistics, Mathematics and Insurance, Assiut University, Egypt. Abstract Three types of regression models researchers need to be familiar with and know the requirements of each: parametric, semiparametric and nonparametric regression models. The type of modeling used is based on how much information are available about the form of the relationship between response variable and explanatory variables, and the random error distribution. In this article, differences between models, common methods of estimation, robust estimation, and applications are introduced. The R code for all the graphs and analyses presented here, in this article, is available in the Appendix. Keywords: Parametric, semiparametric, nonparametric, single index model, robust estimation, kernel regression, spline smoothing. 1 Introduction The aim of this paper is to answer many questions researchers may have when they fit their data, such as what are the differences between parametric, semi/nonparamertic models? which one should be used to model a real data set? what estimation method should be used?, and which type of modeling is better? These questions and others are addressed by examples. arXiv:1906.10221v1 [stat.ME] 24 Jun 2019 Assume that a researcher collected data about Y, a response variable, and explanatory variables, X = (x1; x2; : : : ; xk). The model that describes the relationship between Y and X can be written as Y = f(X; β) + , (1) where β is a vector of k parameters, is an error term whose distribution may or may not be normal, f(·) is some function that describe the relationship between Y and X. -

Bayesian Nonparametric Regression for Educational Research
Bayesian Nonparametric Regression for Educational Research George Karabatsos Professor of Educational Psychology Measurement, Evaluation, Statistics and Assessments University of Illinois-Chicago 2015 Annual Meeting Professional Development Course Thu, April 16, 8:00am to 3:45pm, Swissotel, Event Centre Second Level, Vevey 1&2 Supported by NSF-MMS Research Grant SES-1156372 Session 11.013 - Bayesian Nonparametric Regression for Educational Research Thu, April 16, 8:00am to 3:45pm, Swissotel, Event Centre Second Level, Vevey 1&2 Session Type: Professional Development Course Abstract Regression analysis is ubiquitous in educational research. However, the standard regression models can provide misleading results in data analysis, because they make assumptions that are often violated by real educational data sets. In order to provide an accurate regression analysis of a data set, it is necessary to specify a regression model that can describe all possible patterns of data. The Bayesian nonparametric (BNP) approach provides one possible way to construct such a flexible model, defined by an in…finite- mixture of regression models, that makes minimal assumptions about data. This one-day course will introduce participants to BNP regression models, in a lecture format. The course will also show how to apply the contemporary BNP models, in the analysis of real educational data sets, through guided hands-on exercises. The objective of this course is to show researchers how to use these BNP models for the accurate regression analysis of real educational data, involving either continuous, binary, ordinal, or censored dependent variables; for multi-level analysis, quantile regression analysis, density (distribution) regression analysis, causal analysis, meta-analysis, item response analysis, automatic predictor selection, cluster analysis, and survival analysis.