Download Author Version (PDF)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Pharmacy and Poisons (Third and Fourth Schedule Amendment) Order 2017
Q UO N T FA R U T A F E BERMUDA PHARMACY AND POISONS (THIRD AND FOURTH SCHEDULE AMENDMENT) ORDER 2017 BR 111 / 2017 The Minister responsible for health, in exercise of the power conferred by section 48A(1) of the Pharmacy and Poisons Act 1979, makes the following Order: Citation 1 This Order may be cited as the Pharmacy and Poisons (Third and Fourth Schedule Amendment) Order 2017. Repeals and replaces the Third and Fourth Schedule of the Pharmacy and Poisons Act 1979 2 The Third and Fourth Schedules to the Pharmacy and Poisons Act 1979 are repealed and replaced with— “THIRD SCHEDULE (Sections 25(6); 27(1))) DRUGS OBTAINABLE ONLY ON PRESCRIPTION EXCEPT WHERE SPECIFIED IN THE FOURTH SCHEDULE (PART I AND PART II) Note: The following annotations used in this Schedule have the following meanings: md (maximum dose) i.e. the maximum quantity of the substance contained in the amount of a medicinal product which is recommended to be taken or administered at any one time. 1 PHARMACY AND POISONS (THIRD AND FOURTH SCHEDULE AMENDMENT) ORDER 2017 mdd (maximum daily dose) i.e. the maximum quantity of the substance that is contained in the amount of a medicinal product which is recommended to be taken or administered in any period of 24 hours. mg milligram ms (maximum strength) i.e. either or, if so specified, both of the following: (a) the maximum quantity of the substance by weight or volume that is contained in the dosage unit of a medicinal product; or (b) the maximum percentage of the substance contained in a medicinal product calculated in terms of w/w, w/v, v/w, or v/v, as appropriate. -

Title 16. Crimes and Offenses Chapter 13. Controlled Substances Article 1
TITLE 16. CRIMES AND OFFENSES CHAPTER 13. CONTROLLED SUBSTANCES ARTICLE 1. GENERAL PROVISIONS § 16-13-1. Drug related objects (a) As used in this Code section, the term: (1) "Controlled substance" shall have the same meaning as defined in Article 2 of this chapter, relating to controlled substances. For the purposes of this Code section, the term "controlled substance" shall include marijuana as defined by paragraph (16) of Code Section 16-13-21. (2) "Dangerous drug" shall have the same meaning as defined in Article 3 of this chapter, relating to dangerous drugs. (3) "Drug related object" means any machine, instrument, tool, equipment, contrivance, or device which an average person would reasonably conclude is intended to be used for one or more of the following purposes: (A) To introduce into the human body any dangerous drug or controlled substance under circumstances in violation of the laws of this state; (B) To enhance the effect on the human body of any dangerous drug or controlled substance under circumstances in violation of the laws of this state; (C) To conceal any quantity of any dangerous drug or controlled substance under circumstances in violation of the laws of this state; or (D) To test the strength, effectiveness, or purity of any dangerous drug or controlled substance under circumstances in violation of the laws of this state. (4) "Knowingly" means having general knowledge that a machine, instrument, tool, item of equipment, contrivance, or device is a drug related object or having reasonable grounds to believe that any such object is or may, to an average person, appear to be a drug related object. -

QSAR Model for Androgen Receptor Antagonism
s & H oid orm er o t n S f a l o S l c a Journal of i n e Jensen et al., J Steroids Horm Sci 2012, S:2 r n u c o e DOI: 10.4172/2157-7536.S2-006 J ISSN: 2157-7536 Steroids & Hormonal Science Research Article Open Access QSAR Model for Androgen Receptor Antagonism - Data from CHO Cell Reporter Gene Assays Gunde Egeskov Jensen*, Nikolai Georgiev Nikolov, Karin Dreisig, Anne Marie Vinggaard and Jay Russel Niemelä National Food Institute, Technical University of Denmark, Department of Toxicology and Risk Assessment, Mørkhøj Bygade 19, 2860 Søborg, Denmark Abstract For the development of QSAR models for Androgen Receptor (AR) antagonism, a training set based on reporter gene data from Chinese hamster ovary (CHO) cells was constructed. The training set is composed of data from the literature as well as new data for 51 cardiovascular drugs screened for AR antagonism in our laboratory. The data set represents a wide range of chemical structures and various functions. Twelve percent of the screened drugs were AR antagonisms; three out of six statins showed AR antagonism, two showed cytotoxicity and one was negative. The newly identified AR antagonisms are: Lovastatin, Simvastatin, Mevastatin, Amiodaron, Docosahexaenoic acid and Dilazep. A total of 874 (231 positive, 643 negative) chemicals constitute the training set for the model. The Case Ultra expert system was used to construct the QSAR model. The model was cross-validated (leave-groups-out) with a concordance of 78.4%, a specificity of 86.1% and a sensitivity of 57.9%. -
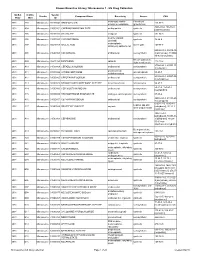
Known Bioactive Library: Microsource 1 - US Drug Collection
Known Bioactive Library: Microsource 1 - US Drug Collection ICCB-L ICCB-L Vendor Vendor Compound Name Bioactivity Source CAS Plate Well ID antifungal, inhibits Penicillium 2091 A03 Microsource 00200046 GRISEOFULVIN 126-07-8 mitosis in metaphase griseofulvum 3505-38-2, 486-16-8 2091 A04 Microsource 01500161 CARBINOXAMINE MALEATE antihistaminic synthetic [carbinoxamine] 2091 A05 Microsource 00200331 SALSALATE analgesic synthetic 552-94-3 muscle relaxant 2091 A06 Microsource 01500162 CARISOPRODOL synthetic 78-44-4 (skeletal) antineoplastic, 2091 A07 Microsource 00210369 GALLIC ACID insect galls 149-91-7 astringent, antibacterial 66592-87-8, 50370-12- 2091 A08 Microsource 01500163 CEFADROXIL antibacterial semisynthetic 2 [anhydrous], 119922- 89-9 [hemihydrate] Rheum palmatum, 2091 A09 Microsource 00211468 DANTHRON cathartic 117-10-2 Xyris semifuscata 27164-46-1, 25953-19- 2091 A10 Microsource 01500164 CEFAZOLIN SODIUM antibacterial semisynthetic 9 [cefazolin] glucocorticoid, 2091 A11 Microsource 00300024 HYDROCORTISONE adrenal glands 50-23-7 antiinflammatory 64485-93-4, 63527-52- 2091 A12 Microsource 01500165 CEFOTAXIME SODIUM antibacterial semisynthetic 6 [cefotaxime] 2091 A13 Microsource 00300029 DESOXYCORTICOSTERONE ACETATE mineralocorticoid adrenocortex 56-47-3 58-71-9, 153-61-7 2091 A14 Microsource 01500166 CEPHALOTHIN SODIUM antibacterial semisynthetic [cephalothin] 2091 A15 Microsource 00300034 TESTOSTERONE PROPIONATE androgen, antineoplastic semisynthetic 57-85-2 24356-60-3, 21593-23- 2091 A16 Microsource 01500167 CEPHAPIRIN SODIUM -

Niacin Alternatives for Dyslipidemia: Fool's Gold Or Gold Mine? Part II
Curr Atheroscler Rep (2016) 18: 17 DOI 10.1007/s11883-016-0570-9 NONSTATIN DRUGS (E. DEGOMA, SECTION EDITOR) Niacin Alternatives for Dyslipidemia: Fool’s Gold or Gold Mine? Part II: Novel Niacin Mimetics Harsh Goel1 & Richard L. Dunbar2,3,4,5,6 Published online: 1 March 2016 # The Author(s) 2016. This article is published with open access at Springerlink.com Abstract Two cardiovascular outcome trials established ni- predecessors. Thus, shrewd developers could effectively acin 3 g daily prevents hard cardiac events. However, as tap into a gold mine at the grave of the ER alternative. The detailed in part I of this series, an extended-release (ER) GPR109A receptor was discovered a decade ago, leading to alternative at only 2 g nightly demonstrated no comparable a large body of evidence commending the niacin pathway to benefits in two outcome trials, implying the alternative is a lower cardiovascular risk beyond statins. While mediating not equivalent to the established cardioprotective regimen. niacin’s most prominent adverse effects, GPR109A also Since statins leave a significant treatment gap, this presents seems to mediate anti-lipolytic, anti-inflammatory, and a major opportunity for developers. Importantly, the anti-atherogenic effects of niacin. Several developers are established regimen is cardioprotective, so the pathway is investing heavily in novel strategies to exploit niacin’sther- likely beneficial. Moreover, though effective, the apeutic pathways. These include selective GPR109A recep- established cardioprotective regimen is cumbersome, limit- tor agonists, niacin prodrugs, and a niacin metabolite, with ing clinical use. At the same time, the ER alternative has encouraging early phase human data. -

2 12/ 35 74Al
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date 22 March 2012 (22.03.2012) 2 12/ 35 74 Al (51) International Patent Classification: (81) Designated States (unless otherwise indicated, for every A61K 9/16 (2006.01) A61K 9/51 (2006.01) kind of national protection available): AE, AG, AL, AM, A61K 9/14 (2006.01) AO, AT, AU, AZ, BA, BB, BG, BH, BR, BW, BY, BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, DO, (21) International Application Number: DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, PCT/EP201 1/065959 HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KM, KN, KP, (22) International Filing Date: KR, KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, 14 September 201 1 (14.09.201 1) ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, PE, PG, PH, PL, PT, QA, RO, RS, RU, (25) Filing Language: English RW, SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, (26) Publication Language: English TM, TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (30) Priority Data: 61/382,653 14 September 2010 (14.09.2010) US (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (71) Applicant (for all designated States except US): GM, KE, LR, LS, MW, MZ, NA, SD, SL, SZ, TZ, UG, NANOLOGICA AB [SE/SE]; P.O Box 8182, S-104 20 ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, MD, RU, TJ, Stockholm (SE). -

(12) Patent Application Publication (10) Pub. No.: US 2004/0053975A1 Bova Et Al
US 2004.0053975A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2004/0053975A1 Bova et al. (43) Pub. Date: Mar. 18, 2004 (54) COMBINATIONS OF HMG-COA tor, which are useful for altering lipid levels in Subjects REDUCTASE INHIBITORS AND NCOTINC Suffering from, for example, hyperlipidemia and atheroscle ACID COMPOUNDS AND METHODS FOR rosis, without causing drug-induced hepatotoxicity, myopa TREATING HYPELPIDEMIA ONCE A DAY thy or rhabdomyolysis. The present invention also relates to AT NIGHT methods of altering Serum lipids in Subjects to treat, for example, hyperlipidemia in hyperlipidemics, lipidemia in (76) Inventors: David J. Bova, Boca Raton, FL (US); normolipidemics diagnosed with or predisposed to cardio Josephine Dunne, Plantation, FL (US) vascular disease, and atherosclerosis, by administering Such Correspondence Address: oral Solid pharmaceutical combinations once per day as a Karen J. Messick Single dose during the evening hours, without causing drug Kos Pharmaceuticals, Inc. induced hepatotoxicity, myopathy or rhabdomyolysis, or 25th Floor without causing in at least an appreciable number of indi 1001 Brickell Bay Drive viduals drug-induced hepatotoxicity, myopathy or rhab Miami, FL 33131 (US) domyolysis to Such a level that discontinuation of Such therapy would be required. More particularly, the present (21) Appl. No.: 10/260,027 invention concerns oral Solid pharmaceutical combinations comprised of, for example, (1) an HMG-CoA reductase (22) Filed: Sep. 2, 2003 inhibitor for immediate or extended release, (2) nicotinic acid, a nicotinic acid compound or mixtures thereof and (3) Related U.S. Application Data a Swelling agent to form a Sustained release composition for extended release of the nicotinic acid or nicotinic acid (63) Continuation of application No. -

Trends in Lipid-Modifying Agent Use in 83 Countries
medRxiv preprint doi: https://doi.org/10.1101/2021.01.10.21249523; this version posted January 11, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. All rights reserved. No reuse allowed without permission. Trends in lipid-modifying agent use in 83 countries Authors (ORCID) Joseph E Blais (0000-0001-7895-198X)1; Yue Wei (NA)1; Kevin KW Yap (NA)2; Hassan Alwafi (NA)3; Tian-Tian Ma (0000-0003-1361-4055)3; Ruth Brauer (0000- 0001-8934-347X)3; Wallis CY Lau (0000-0003-2320-0470)3; Kenneth KC Man (0000- 0001-8645-1942)3; Chung Wah Siu (0000-0002-5570-983X)4; Kathryn C Tan (0000- 0001-9037-0416)5; Ian CK Wong (0000-0001-8242-0014)1,3; Li Wei (0000-0001- 8840-7267)3; Esther W Chan (0000-0002-7602-9470)1 Affiliations 1 Centre for Safe Medication Practice and Research, Department of Pharmacology and Pharmacy, LKS Faculty of Medicine, University of Hong Kong, Hong Kong Special Administrative Region, China 2 Department of Pharmacology and Pharmacy, LKS Faculty of Medicine, University of Hong Kong, Hong Kong Special Administrative Region, China 3 Research Department of Practice and Policy, UCL School of Pharmacy, London, UK 4 Division of Cardiology, Department of Medicine, University of Hong Kong, Hong Kong Special Administrative Region, China 5 Department of Medicine, University of Hong Kong, Hong Kong Special Administrative Region, China Correspondence Esther W Chan, PhD Centre for Safe Medication Practice and Research Department of Pharmacology and Pharmacy NOTE: This preprint reports new research that has not been certified by peer review and should not be used to guide clinical practice. -

Under-Utilisation of Preventive Medication in Patients with Cardiovascular Disease Is Greatest in Younger Age Groups (PREDICT-CVD 15)
ORIGINAL SCIENTIFIC PAPERS QUANTItatIVE RESEARCH Under-utilisation of preventive medication in patients with cardiovascular disease is greatest in younger age groups (PREDICT-CVD 15) Suneela Mehta MBChB, MPH;1 Sue Wells MBChB, MPH, PhD;1 Tania Riddell BSc, MBChB, MPH;1 Andrew 1 Section of Epidemiology Kerr MA, MBChB;2 Romana Pylypchuk MSc, MPH;1 Roger Marshall BSc, MSc, PhD;1 Shanthi Ameratunga and Biostatistics, School of Population Health, The 1 3 1 MBChB, MPH, PhD; Wing Cheuk Chan MBChB, MPH; Simon Thornley MBChB, MPH; Sue Crengle University of Auckland, MBChB, MPH, PhD;4 Jeff Harrison BSc (Hons), PhD;5 Paul Drury MA MB BChir;6 C Raina Elley MBChB, PhD;7 Auckland, New Zealand Fionna Bell MBChB, MPH;8 Rod Jackson MBChB, PhD1 2 Middlemore Hospital, Counties Manukau District Health Board, Auckland ABSTRACT 3 Health Intelligence Unit, Counties Manukau District INtroductioN: Blood pressure–lowering (BPL) and lipid-lowering (LL) medications together reduce Health Board, Auckland estimated absolute five-year cardiovascular disease (CVD) risk by >40%. International studies indicate 4 that the proportion of people with CVD receiving pharmacotherapy increases with advancing age. Te Kupenga Hauora Maori, School of Population Health, Aim: To compare BPL and LL medications, by sociodemographic characteristics, for patients with known The University of Auckland, Auckland CVD in primary care settings. 5 Division of MethodS: The study population included patients aged 35–74 with known CVD assessed in primary Pharmacotherapy, School of care from July 2006 to October 2009 using a web-based computerised decision support system (PRE- Pharmacy, The University of DICT) for risk assessment and management. -

(12) United States Patent (Lo) Patent No.: �US 8,480,637 B2
111111111111111111111111111111111111111111111111111111111111111111111111 (12) United States Patent (lo) Patent No.: US 8,480,637 B2 Ferrari et al. (45) Date of Patent : Jul. 9, 2013 (54) NANOCHANNELED DEVICE AND RELATED USPC .................. 604/264; 907/700, 902, 904, 906 METHODS See application file for complete search history. (75) Inventors: Mauro Ferrari, Houston, TX (US); (56) References Cited Xuewu Liu, Sugar Land, TX (US); Alessandro Grattoni, Houston, TX U.S. PATENT DOCUMENTS (US); Daniel Fine, Austin, TX (US); 5,651,900 A 7/1997 Keller et al . .................... 216/56 Randy Goodall, Austin, TX (US); 5,728,396 A 3/1998 Peery et al . ................... 424/422 Sharath Hosali, Austin, TX (US); Ryan 5,770,076 A 6/1998 Chu et al ....................... 210/490 5,798,042 A 8/1998 Chu et al ....................... 210/490 Medema, Pflugerville, TX (US); Lee 5,893,974 A 4/1999 Keller et al . .................. 510/483 Hudson, Elgin, TX (US) 5,938,923 A 8/1999 Tu et al . ........................ 210/490 5,948,255 A * 9/1999 Keller et al . ............. 210/321.84 (73) Assignees: The Board of Regents of the University 5,985,164 A 11/1999 Chu et al ......................... 516/41 of Texas System, Austin, TX (US); The 5,985,328 A 11/1999 Chu et al ....................... 424/489 Ohio State University Research (Continued) Foundation, Columbus, OH (US) FOREIGN PATENT DOCUMENTS (*) Notice: Subject to any disclaimer, the term of this WO WO 2004/036623 4/2004 WO WO 2006/113860 10/2006 patent is extended or adjusted under 35 WO WO 2009/149362 12/2009 U.S.C. 154(b) by 612 days. -

(12) United States Patent (Io) Patent No.: US 9,526,824 B2 Ferrari Et Al
1111111111111111111111111111111111111111111111111111111111111111111111 (12) United States Patent (io) Patent No.: US 9,526,824 B2 Ferrari et al. (45) Date of Patent: Dec. 27, 2016 (54) NANOCHANNELED DEVICE AND RELATED (52) U.S. Cl. METHODS CPC .............. A61M 5/00 (2013.01); A61K 9/0097 (2013.01); A61M37/00 (2013.01); (71) Applicants:The Board of Regents of the (Continued) University of Texas System, Austin, TX (US); The Ohio State University (58) Field of Classification Search Research Foundation, Columbus, OH CPC ............. B81C 1/00444; B81C 1/00476; B81C (US) 1/00119; BO1D 67/0034; YIOS 148/05 (Continued) (72) Inventors: Mauro Ferrari, Houston, TX (US); Xuewu Liu, Sugar Land, TX (US); (56) References Cited Alessandro Grattoni, Houston, TX (US); Daniel Fine, Austin, TX (US); U.S. PATENT DOCUMENTS Randy Goodall, Austin, TX (US); Sharath Hosali, Austin, TX (US); 3,731,681 A 5/1973 Blackshear et al. Ryan Medema, Pflugerville, TX (US); 3,921,636 A 11/1975 Zaffaroni Lee Hudson, Elgin, TX (US) (Continued) (73) Assignees: The Board of Regents of the FOREIGN PATENT DOCUMENTS University of Texas System, Austin, TX (US); The Ohio State University CN 1585627 2/2005 Research Foundation, Columbus, OH DE 10 2006 014476 7/2007 (US) (Continued) (*) Notice: Subject to any disclaimer, the term of this OTHER PUBLICATIONS patent is extended or adjusted under 35 U.S.C. 154(b) by 784 days. "The Economic Costs of Drug Abuse in the United States," www. whitehousedrugpolicy.gov, Sep. 2001. (21) Appl. No.: 13/875,871 (Continued) (22) Filed: May 2, 2013 Primary Examiner Binh X Tran (65) Prior Publication Data (74) Attorney, Agent, or Firm Parker Highlander PLLC US 2013/0240483 Al Sep. -

Medicines Amendment Regulations (No 2) 2015
Reprint as at 25 October 2018 Medicines Amendment Regulations (No 2) 2015 (LI 2015/180) Medicines Amendment Regulations (No 2) 2015: revoked, on 25 October 2018, by regulation 5 of the Medicines Amendment Regulations 2018 (LI 2018/179). Jerry Mateparae, Governor-General Order in Council At Wellington this 27th day of July 2015 Present: His Excellency the Governor-General in Council Pursuant to section 105(1)(j) of the Medicines Act 1981, His Excellency the Governor-General makes the following regulations, acting— (a) on the advice and with the consent of the Executive Council; and (b) on the advice of the Minister of Health tendered after consultation with the organisations or bodies appearing to the Minister to be representative of per- sons likely to be substantially affected. Contents Page 1 Title 2 2 Commencement 2 3 Principal regulations 2 4 Schedule 1 replaced 2 Note Changes authorised by subpart 2 of Part 2 of the Legislation Act 2012 have been made in this official reprint. Note 4 at the end of this reprint provides a list of the amendments incorporated. These regulations are administered by the Ministry of Health. 1 Reprinted as at r 1 Medicines Amendment Regulations (No 2) 2015 25 October 2018 Schedule 2 Schedule 1 replaced Regulations 1 Title These regulations are the Medicines Amendment Regulations (No 2) 2015. 2 Commencement These regulations come into force on 6 August 2015. 3 Principal regulations These regulations amend the Medicines Regulations 1984 (the principal regu- lations). 4 Schedule 1 replaced Replace Schedule 1 with the Schedule 1 set out in the Schedule of these regula- tions.