Metabolic Programming a Lean Phenotype by Deregulation of RNA Polymerase III
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Development of a System of High Ornithine and Citrulline Production by a Plant-Derived Lactic Acid Bacterium, Weissella Confusa K-28
Development of a system of high ornithine and citrulline production by a plant-derived lactic acid bacterium, Weissella confusa K-28 㸦᳜≀⏤᮶ங㓟⳦ :HLVVHOODFRQIXVD. ࠾ࡅࡿ ࢜ࣝࢽࢳࣥཬࡧࢩࢺࣝࣜࣥ㧗⏕⏘ࢩࢫࢸ࣒ࡢᵓ⠏㸧 Md Rakhimuzzaman Student ID: D164180 Supervisor: Prof. Masanori Sugiyama Department of Probiotic Science for Preventive Medicine Graduate School of Biomedical and Health Sciences Hiroshima University, Japan 2019 ABBREVIATIONS ADI arginine deiminase AUC area under curve BLAST basic local alignment search tool CK carbamate kinase dNTP deoxyribonucleotide triphosphate DDBJ DNA Data Bank of Japan EDTA ethylenediamine tetra-acetic acid EtOH ethanol EPS exopolysaccharide GABA gamma-aminobutyric acid GRAS generally recognized as safe HPLC high performance liquid chromatography LAB lactic acid bacteria Lb. Lactobacillus OD optical density OTC ornithine carbamoyltransferase PITC phenyl isothiocyanate TAE tris-acetate-ethylenediamine tetra-acetic acid TE tris-ethylenediamine tetra-acetic acid TEA triethylamine Tris tris(hydroxymethyl)aminomethane rDNA ribosomal deoxyribonucleic acid RT-PCR reverse transcription polymerase chain reaction CONTENTS INTRODUCTION 1-17 OBJECTIVES 18 MATERIALS AND METHODS 19-34 RESULTS 35-57 DISCUSSIONS 58-65 CONCLUSION 66-67 ACKNOWLEDGEMENTS 68 REFERENCES 69-84 INTRODUCTION Lactic acid bacteria (LAB) are an order of gram-positive, acid-tolerant, generally nonsporulating, non-respiring, either rod-shaped or spherical microorganisms. Since LAB lack the ability of synthesizing porphyrins (heme, and components of respiratory chains) (Pessione et al, 2010), the bacteria can not generate ATP without external heme supplementation (Pessione et al, 2010). The LAB strains can only obtain ATP by fermentation, usually they generate the ATP from some sugars. Because LAB do not use oxygen for energy production, the bacteria easily grow under anaerobic conditions, but they can also grow in presence of oxygen. -

Metabolic Reprogramming in Prostate Cancer
www.nature.com/bjc REVIEW ARTICLE Metabolic reprogramming in prostate cancer Fahim Ahmad 1,2, Murali Krishna Cherukuri2 and Peter L. Choyke1 Although low risk localised prostate cancer has an excellent prognosis owing to effective treatments, such as surgery, radiation, cryosurgery and hormone therapy, metastatic prostate cancer remains incurable. Existing therapeutic regimens prolong life; however, they are beset by problems of resistance, resulting in poor outcomes. Treatment resistance arises primarily from tumour heterogeneity, altered genetic signatures and metabolic reprogramming, all of which enable the tumour to serially adapt to drugs during the course of treatment. In this review, we focus on alterations in the metabolism of prostate cancer, including genetic signatures and molecular pathways associated with metabolic reprogramming. Advances in our understanding of prostate cancer metabolism might help to explain many of the adaptive responses that are induced by therapy, which might, in turn, lead to the attainment of more durable therapeutic responses. British Journal of Cancer https://doi.org/10.1038/s41416-021-01435-5 BACKGROUND Several tools have been designed to assess metabolism without Cellular metabolism involves complex biochemical processes disturbing the system. Advances in nuclear magnetic resonance through which specific nutrients are consumed. Carbohydrates, and mass spectroscopy have helped to quantify the carbon influx fatty acids and amino acids are the main source of nutrients for of the central metabolic pathways (e.g. TCA cycle, glycolysis and energy homeostasis and macromolecular synthesis. They are the pentose phosphate pathway) in mammalian systems, and this main constituent of core metabolic pathways that can be approach has been instrumental in demonstrating that the classified as anabolic, catabolic or waste producing (Fig. -

35 Disorders of Purine and Pyrimidine Metabolism
35 Disorders of Purine and Pyrimidine Metabolism Georges van den Berghe, M.- Françoise Vincent, Sandrine Marie 35.1 Inborn Errors of Purine Metabolism – 435 35.1.1 Phosphoribosyl Pyrophosphate Synthetase Superactivity – 435 35.1.2 Adenylosuccinase Deficiency – 436 35.1.3 AICA-Ribosiduria – 437 35.1.4 Muscle AMP Deaminase Deficiency – 437 35.1.5 Adenosine Deaminase Deficiency – 438 35.1.6 Adenosine Deaminase Superactivity – 439 35.1.7 Purine Nucleoside Phosphorylase Deficiency – 440 35.1.8 Xanthine Oxidase Deficiency – 440 35.1.9 Hypoxanthine-Guanine Phosphoribosyltransferase Deficiency – 441 35.1.10 Adenine Phosphoribosyltransferase Deficiency – 442 35.1.11 Deoxyguanosine Kinase Deficiency – 442 35.2 Inborn Errors of Pyrimidine Metabolism – 445 35.2.1 UMP Synthase Deficiency (Hereditary Orotic Aciduria) – 445 35.2.2 Dihydropyrimidine Dehydrogenase Deficiency – 445 35.2.3 Dihydropyrimidinase Deficiency – 446 35.2.4 Ureidopropionase Deficiency – 446 35.2.5 Pyrimidine 5’-Nucleotidase Deficiency – 446 35.2.6 Cytosolic 5’-Nucleotidase Superactivity – 447 35.2.7 Thymidine Phosphorylase Deficiency – 447 35.2.8 Thymidine Kinase Deficiency – 447 References – 447 434 Chapter 35 · Disorders of Purine and Pyrimidine Metabolism Purine Metabolism Purine nucleotides are essential cellular constituents 4 The catabolic pathway starts from GMP, IMP and which intervene in energy transfer, metabolic regula- AMP, and produces uric acid, a poorly soluble tion, and synthesis of DNA and RNA. Purine metabo- compound, which tends to crystallize once its lism can be divided into three pathways: plasma concentration surpasses 6.5–7 mg/dl (0.38– 4 The biosynthetic pathway, often termed de novo, 0.47 mmol/l). starts with the formation of phosphoribosyl pyro- 4 The salvage pathway utilizes the purine bases, gua- phosphate (PRPP) and leads to the synthesis of nine, hypoxanthine and adenine, which are pro- inosine monophosphate (IMP). -

Amino Acid Degradation; Urea Cycle
Amino acid degradation; urea cycle Kristina Mlinac Jerković [email protected] In animals, amino acids undergo oxidative degradation in three different metabolic circumstances: 1. During the normal synthesis and degradation of cellular proteins, some amino acids that are released from protein breakdown and are not needed for new protein synthesis undergo oxidative degradation. 2. When a diet is rich in protein and the ingested amino acids exceed the body’s needs for protein synthesis, the surplus is catabolized; amino acids cannot be stored. 3. During starvation or in uncontrolled diabetes mellitus, when carbohydrates are either unavailable or not properly utilized, cellular proteins are used as fuel. Metabolic fates of amino groups - The metabolism of nitrogen-containing molecules such as proteins and nucleic acids differs significantly from that of carbohydrates and lipids. - Whereas the latter molecules can be stored and mobilized as needed for biosynthetic reactions or for energy generation, there is no nitrogen-storing molecule (one exception to this rule is storage protein in seeds). - Organisms must constantly replenish their supply of usable nitrogen to replace organic nitrogen that is lost in catabolism. For example, animals must have a steady supply of amino acids in their diets to replace the nitrogen excreted as urea, uric acid, and other nitrogenous waste products. - Since excess dietary amino acids are neither stored for future use nor excreted, they are converted to common metabolic intermediates such as pyruvate, oxaloacetate, acetyl-CoA, and α-keto-glutarate. - Consequently, amino acids are also precursors of glucose, fatty acids, and ketone bodies and are therefore metabolic fuels. -

Protein Metabolism and It's Disorders
Protein Metabolism and it’s Disorders (Course : M.Sc. Zoology) Course Code: ZOOL 4008 (Biochemistry and Metabolism), Semester –II Dr. Shyam Babu Prasad Assistant Professor Department of Zoology Mahatma Gandhi Central University (MGCU), Motihari-845401 (Bihar) Email: [email protected] Important facts of the Protein Metabolism •Proteins metabolism referred as synthesis of protein from Amino acids (Anabolism) and breakdown of proteins (Catabolism). •In the Unit II, we discussed the synthesis (Anabolism) of various form of the proteins from different amino acids. • Here we mainly focused on the Catabolism of the protein, which is integral part of metabolism of the nitrogen-containing molecules. • Nitrogen enters in to the body in various form of compounds that is present in food, the most important is amino acids (as dietary food). • During Protein/amino acid metabolism (Catabolism), Nitrogen leaves the body as urea, ammonia, and in form of other derivatives. • The Catabolism of the protein depends on the amino acid pool and protein turnover (the rate of protein synthesis and degradation is equal). • The amino acids present in through out of the body (cells, blood, and the extracellular fluids etc) is called amino acid pools. It is maintained by following factors as represented in below diagram: Body protein Dietary degradation Protein 400gm/day 600gm/day Supplied Amino acid pool Depleted Synthesis of Remaining Synthesis of Glucose, Synthesis of amino acids Nitrogen containing Glycogen, the Body burned as compound like Ketone Proteins -
Transaminations and Urea Cycle
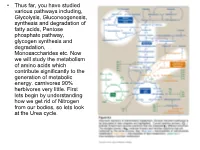
• Thus far, you have studied various pathways including, Glycolysis, Gluconeogenesis, synthesis and degradation of fatty acids, Pentose phosphate pathway, glycogen synthesis and degradation, Monosaccharides etc. Now we will study the metabolism of amino acids which contribute significantly to the generation of metabolic energy. carnivores 90% herbivores very little. First lets begin by understanding how we get rid of Nitrogen from our bodies, so lets look at the Urea cycle. 1 α Amino group keeps amino acids safe from oxidative breakdown. Removal of this group is essential for energy production it is an obligatory step for catabolism. Nitrogen can then be incorporated or excreted.This is done through transamination or oxidative deamination, provide amonia and aspartate sources for the Urea cycle. Transamination is funneled through glutamate. This is done through the transfer of the α amino group to α ketoglutarate. Product α-keto acid and glutamate. Alanine to pyruvate and aspartate to oxaloacetate all amino acids except lysine and threonina (lose the α amino through deamination. Glutamate dehydrogenase the oxidative deamination Glutamate only amino acid that undergoes rapid oxidative deamination (glutamate dehydrogenase NH3 + NADH), where amino groups are released as ammonia. After a meal rich in protein the levels of glutamate in liver are increased favoring amino acid degradation and formation of ammonia. Activators of this enzyme are ADP and GDP while ATP and GTP are inhibitors. This reaction occurs in the mitochondria. Ammonia can be transported to the liver from other tissues through two mechanisms that reduce the toxicity of ammonia. One through Glutamine (most tissues through, glutamine synthetase). In the liver the glutamine is converted back to glutamate (Fig. -

1 T CELL ACTIVATION TRIGGERS REVERSIBLE INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE ASSEMBLY Krisna C. Duong-Ly 1, Yin-Ming Kuo 2
bioRxiv preprint doi: https://doi.org/10.1101/315929; this version posted May 7, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. T CELL ACTIVATION TRIGGERS REVERSIBLE INOSINE-5’-MONOPHOSPHATE DEHYDROGENASE ASSEMBLY Krisna C. Duong-Ly1, Yin-Ming Kuo2, Matthew C. Johnson3, Justin M. Kollman3, Jonathan Soboloff4, Glenn F. Rall5, Andrew J. Andrews2, and Jeffrey R. Peterson1 1Cancer Biology Program, Fox Chase Cancer Center, Philadelphia, PA 2Cancer Epigenetics Program, Fox Chase Cancer Center, Philadelphia, PA 3Department of Biochemistry, University of Washington, Seattle, WA 4Fels Institute for Cancer Research and Molecular Biology, Temple University School of Medicine, Philadelphia, PA 5Blood Cell Development and Function Program, Fox Chase Cancer Center, Philadelphia, PA Correspondence to Jeffrey R. Peterson: [email protected], Fox Chase Cancer Center, 333 Cottman Avenue, Philadelphia, PA 19111, ORCID: 0000-0002-0604-718X; Yin-Ming Kuo’s present address is Department of Radiology, University of Pennsylvania School of Medicine, Philadelphia, PA Short running title: IMPDH FILAMENT ASSEMBLY IN T CELLS Abbreviations used: IMP: inosine-5’-monophosphate IMPDH: inosine-5’-monophosphate dehydrogenase 1 bioRxiv preprint doi: https://doi.org/10.1101/315929; this version posted May 7, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. -

Supplementary Table S1 List of Proteins Identified with LC-MS/MS in the Exudates of Ustilaginoidea Virens Mol
Supplementary Table S1 List of proteins identified with LC-MS/MS in the exudates of Ustilaginoidea virens Mol. weight NO a Protein IDs b Protein names c Score d Cov f MS/MS Peptide sequence g [kDa] e Succinate dehydrogenase [ubiquinone] 1 KDB17818.1 6.282 30.486 4.1 TGPMILDALVR iron-sulfur subunit, mitochondrial 2 KDB18023.1 3-ketoacyl-CoA thiolase, peroxisomal 6.2998 43.626 2.1 ALDLAGISR 3 KDB12646.1 ATP phosphoribosyltransferase 25.709 34.047 17.6 AIDTVVQSTAVLVQSR EIALVMDELSR SSTNTDMVDLIASR VGASDILVLDIHNTR 4 KDB11684.1 Bifunctional purine biosynthetic protein ADE1 22.54 86.534 4.5 GLAHITGGGLIENVPR SLLPVLGEIK TVGESLLTPTR 5 KDB16707.1 Proteasomal ubiquitin receptor ADRM1 12.204 42.367 4.3 GSGSGGAGPDATGGDVR 6 KDB15928.1 Cytochrome b2, mitochondrial 34.9 58.379 9.4 EFDPVHPSDTLR GVQTVEDVLR MLTGADVAQHSDAK SGIEVLAETMPVLR 7 KDB12275.1 Aspartate 1-decarboxylase 11.724 112.62 3.6 GLILTLSEIPEASK TAAIAGLGSGNIIGIPVDNAAR 8 KDB15972.1 Glucosidase 2 subunit beta 7.3902 64.984 3.2 IDPLSPQQLLPASGLAPGR AAGLALGALDDRPLDGR AIPIEVLPLAAPDVLAR AVDDHLLPSYR GGGACLLQEK 9 KDB15004.1 Ribose-5-phosphate isomerase 70.089 32.491 32.6 GPAFHAR KLIAVADSR LIAVADSR MTFFPTGSQSK YVGIGSGSTVVHVVDAIASK 10 KDB18474.1 D-arabinitol dehydrogenase 1 19.425 25.025 19.2 ENPEAQFDQLKK ILEDAIHYVR NLNWVDATLLEPASCACHGLEK 11 KDB18473.1 D-arabinitol dehydrogenase 1 11.481 10.294 36.6 FPLIPGHETVGVIAAVGK VAADNSELCNECFYCR 12 KDB15780.1 Cyanovirin-N homolog 85.42 11.188 31.7 QVINLDER TASNVQLQGSQLTAELATLSGEPR GAATAAHEAYK IELELEK KEEGDSTEKPAEETK LGGELTVDER NATDVAQTDLTPTHPIR 13 KDB14501.1 14-3-3 -

Nucleo'de)Metabolism) (Chapter)23)) INTRODUCTION)TO)NUCLEIC)ACIDS)
BIOCH 765: Biochemistry II Spring 2014 Nucleo'de)Metabolism) (Chapter)23)) Jianhan)Chen) Office)Hour:)MF)1:30B2:30PM,)Chalmers)034) Email:)[email protected]) Office:)785B2518) INTRODUCTION)TO)NUCLEIC)ACIDS) (c))Jianhan)Chen) 2) Eukaryo'c)cell) 3 Genome)is)believed)to)define)a)species) 4 Nucleic)Acids) • Polymer(of( nucleotides:(highly( flexible((compared( to(peptides)( • Nucleic(acids(are( universal(in(living( things,(as(they(are( found(in(all(cells( and(viruses.( • Nucleic(acids(were( first(discovered(by( Friedrich(Miescher( in(1871.( (c))Jianhan)Chen) 5 Building)Blocks)of)Nucleo'des) • Phosphate(group(+(pentose(carbohydrate(+( base((nitrogenGcontaining(heterocyclic(ring)( • Deoxyribonucleic(acid((DNA):(A/G/C/T( • Ribonuclei(acid((RNA):(A/G/C/U( Nucleotide nucleobase 6 Nucleo'des)have)many)roles) • Building(blocks(of(the(nucleic(acid(polymers(RNA(and( DNA.( • Energy(transfer(or(energy(coupling(to(drive(biosynthesis( and(other(processes((muscle(contraction,(transport,(etc).( • Oxidation(reduction(reactions.( • Intracellular(signaling.( ATP 7 Pentose)sugars) Base Phosphate sugar • The(pentose(sugar(in(the(nucloetide(is(either(ribose(or( deoxyribose.( ( – The(base(is(added(at(the(1(position(and(phosphates(are(added(at( the(5(position.( – Most(nucleotides,(including(those(incorporated(into(RNA,(contain( ribose.( – 2’GOH(reduced(to(–H(in(deoxynucleotides( – 3’GOH(participates(in(forming(phosphodiester(linkage( – Deoxynucleotides(are(exclusively(used(for(DNA(synthesis.( 8 Nucleo'de)forma'on) O ║ H N The base is added at the 1 6 N position and phosphates -

Metabolic Adaptation and Maladaptation in Adipose Tissue
REVIEW ARTICLE https://doi.org/10.1038/s42255-018-0021-8 Metabolic adaptation and maladaptation in adipose tissue Edward T. Chouchani1,2* and Shingo Kajimura 3,4,5* Adipose tissue possesses the remarkable capacity to control its size and function in response to a variety of internal and exter- nal cues, such as nutritional status and temperature. The regulatory circuits of fuel storage and oxidation in white adipocytes and thermogenic adipocytes (brown and beige adipocytes) play a central role in systemic energy homeostasis, whereas dysreg- ulation of the pathways is closely associated with metabolic disorders and adipose tissue malfunction, including obesity, insulin resistance, chronic inflammation, mitochondrial dysfunction, and fibrosis. Recent studies have uncovered new regulatory ele- ments that control the above parameters and provide new mechanistic opportunities to reprogram fat cell fate and function. In this Review, we provide an overview of the current understanding of adipocyte metabolism in physiology and disease and also discuss possible strategies to alter fuel utilization in fat cells to improve metabolic health. he human body has the remarkable ability to adapt to inter- dysfunction9,10. Dysregulation of mitochondrial biogenesis and nal and external changes, including nutritional status, envi- oxidative phosphorylation (OXPHOS), excess accumulation of Tronmental temperature, infection, circadian rhythm, ageing, extracellular matrix (ECM), and altered adipokines and lipid pro- and more. These adaptations involve dynamic reprogramming of file are strongly associated with adipose tissue malfunction and cellular metabolism in the peripheral tissues. For example, caloric metabolic disorders. Here we review the current understanding of restriction promotes mitochondrial biogenesis and fatty acid oxi- adipocyte metabolism and discuss strategies to reprogram fat cell dation, thereby shifting cellular metabolism preferentially toward fate and metabolism. -

DRAFT Guidance for the Preparation of Toxicological Profiles
DRAFT Guidance for the Preparation of Toxicological Profiles Agency for Toxic Substances and Disease Registry Department of Health and Human Services April 2018 GUIDANCE FOR THE PREPARATION OF TOXICOLOGICAL PROFILES ii CONTENTS CONTENTS .................................................................................................................................................. ii GENERAL ................................................................................................................................................... vi PURPOSE AND SCOPE OF ATSDR TOXICOLOGICAL PROFILES .................................................. xiv QUALITY CRITERIA FOR ANIMAL AND HUMAN STUDIES .......................................................... xvi GENERAL GUIDANCE FOR PREPARING A TOXICOLOGICAL PROFILE ................................... xviii FRONT MATTER ..................................................................................................................................... xxi CHAPTER 1. RELEVANCE TO PUBLIC HEALTH ................................................................................ 1 1.1 OVERVIEW AND U.S. EXPOSURES ........................................................................................ 2 1.2 SUMMARY OF HEALTH EFFECTS .......................................................................................... 3 1.3 MINIMAL RISK LEVELS (MRLS) ............................................................................................. 4 CHAPTER 2. HEALTH EFFECTS ............................................................................................................ -

Thermogenesis in Adipose Tissue Activated by Thyroid Hormone
International Journal of Molecular Sciences Review Thermogenesis in Adipose Tissue Activated by Thyroid Hormone Winifred W. Yau 1 and Paul M. Yen 1,2,* 1 Laboratory of Hormonal Regulation, Cardiovascular and Metabolic Disorders Program, Duke NUS Medical School, Singapore 169857, Singapore; [email protected] 2 Duke Molecular Physiology Institute, Duke University, Durham, NC 27708, USA * Correspondence: [email protected]; Tel.: +65-6516-7666 Received: 23 March 2020; Accepted: 22 April 2020; Published: 24 April 2020 Abstract: Thermogenesis is the production of heat that occurs in all warm-blooded animals. During cold exposure, there is obligatory thermogenesis derived from body metabolism as well as adaptive thermogenesis through shivering and non-shivering mechanisms. The latter mainly occurs in brown adipose tissue (BAT) and muscle; however, white adipose tissue (WAT) also can undergo browning via adrenergic stimulation to acquire thermogenic potential. Thyroid hormone (TH) also exerts profound effects on thermoregulation, as decreased body temperature and increased body temperature occur during hypothyroidism and hyperthyroidism, respectively. We have termed the TH-mediated thermogenesis under thermoneutral conditions “activated” thermogenesis. TH acts on the brown and/or white adipose tissues to induce uncoupled respiration through the induction of the uncoupling protein (Ucp1) to generate heat. TH acts centrally to activate the BAT and browning through the sympathetic nervous system. However, recent studies also show that TH acts peripherally on the BAT to directly stimulate Ucp1 expression and thermogenesis through an autophagy-dependent mechanism. Additionally, THs can exert Ucp1-independent effects on thermogenesis, most likely through activation of exothermic metabolic pathways. This review summarizes thermogenic effects of THs on adipose tissues.