BIOINFORMATICS APPLICATIONS NOTE Doi:10.1093/Bioinformatics/Bts366
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Pathways and Networks Biological Meaning of the Gene Sets
Pathways and Networks Biological meaning of the gene sets Gene ontology terms ? Pathway mapping Linking to Pubmed abstracts or associted MESH terms Regulation by the same transcription factor (module) Protein families and domains Gene set enrichment analysis Over representation analysis 1 Gene set enrichment analysis 1. Given an a priori defined set of genes S. 2. Rank genes (e.g. by t‐value between 2 groups of microarray samples) ranked gene list L. 3. Calculation of an enrichment score (ES) that reflects the degree to which a set S is overrepresented at the extremes (top or bottom) of the entire ranked list L. 4. Estimation the statistical significance (nominal P value) of the ES by using an empirical phenotype‐based permutation test procedure. 5. Adjustment for multiple hypothesis testing by controlling the false discovery rate (FDR). Gene set enrichment analysis Subramanian A et al. Proc Natl Acad Sci (2005) 2 Biochemical and Metabolic Pathways Böhringer Mannheim Signaling networks (in cancer cell) Hannah, Weinberg, Cell. 2000 137 NCI curated pathway maps (http://pid.nci.nih.gov) Signal Transduction Knowledge Environment http://stke.sciencemag.org/cm/ 3 Pathways • Pathways are available mechanistic information •Kyoto Encylopedia of Genes and Genomes, KEGG (http://www.genome.jp/kegg/) BioCYc • EcoCYC Metabolic Pathway Database (E. Coli) •Also for other organisms (e.g. HumanCYC) 4 Pathways • Pathways from Biocarta • http://www.biocarta.com/genes/index.asp Transpath Part of larger BioBase package (commercial) • PathwayBuilder package -
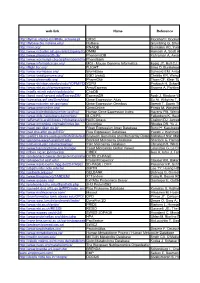
Web-Link Name Reference DRSC Flockhart I, Booker M, Kiger A, Et Al.: Flyrnai: the Drosophila Rnai Screening Center Database
web-link Name Reference http://flyrnai.org/cgi-bin/RNAi_screens.pl DRSC Flockhart I, Booker M, Kiger A, et al.: FlyRNAi: the Drosophila RNAi screening center database. Nucleic Acids Res. 34(Database issue):D489-94 (2006) http://flybase.bio.indiana.edu/ FlyBase Grumbling G, Strelets V.: FlyBase: anatomical data, images and queries. Nucleic Acids Res. 34(Database issue):D484-8 (2006) http://rnai.org/ RNAiDB Gunsalus KC, Yueh WC, MacMenamin P, et al.: RNAiDB and PhenoBlast: web tools for genome-wide phenotypic mapping projects. Nucleic Acids Res. 32(Database issue):D406-10 (2004) http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIMOMIM Hamosh A, Scott AF, Amberger JS, et al.: Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 33(Database issue):D514-7 (2005) http://www.phenomicdb.de/ PhenomicDB Kahraman A, Avramov A, Nashev LG, et al.: PhenomicDB: a multi-species genotype/phenotype database for comparative phenomics. Bioinformatics 21(3):418-20 (2005) http://www.worm.mpi-cbg.de/phenobank2/cgi-bin/MenuPage.pyPhenoBank http://www.informatics.jax.org/ MGI - Mouse Genome Informatics Eppig JT, Bult CJ, Kadin JA, et al.: The Mouse Genome Database (MGD): from genes to mice--a community resource for mouse biology. Nucleic Acids Res. 33(Database issue):D471-5 (2005) http://flight.licr.org/ FLIGHT Sims D, Bursteinas B, Gao Q, et al.: FLIGHT: database and tools for the integration and cross-correlation of large-scale RNAi phenotypic datasets. Nucleic Acids Res. 34(Database issue):D479-83 (2006) http://www.wormbase.org/ WormBase Schwarz EM, Antoshechkin I, Bastiani C, et al.: WormBase: better software, richer content. -

Genmapp 2: New Features and Resources for Pathway Analysis Nathan Salomonis Gladstone Institute of Cardiovascular Disease
Washington University School of Medicine Digital Commons@Becker Open Access Publications 2007 GenMAPP 2: New features and resources for pathway analysis Nathan Salomonis Gladstone Institute of Cardiovascular Disease Kristina Hanspers Gladstone Institute of Cardiovascular Disease Alexander C. Zambon Gladstone Institute of Cardiovascular Disease Karen Vranizan Gladstone Institute of Cardiovascular Disease Steven C. Lawlor Gladstone Institute of Cardiovascular Disease See next page for additional authors Follow this and additional works at: https://digitalcommons.wustl.edu/open_access_pubs Part of the Medicine and Health Sciences Commons Recommended Citation Salomonis, Nathan; Hanspers, Kristina; Zambon, Alexander C.; Vranizan, Karen; Lawlor, Steven C.; Dahlquist, Kam D.; Doniger, Scott .;W Stuart, Josh; Conklin, Bruce R.; and Pico, Alexander R., ,"GenMAPP 2: New features and resources for pathway analysis." BMC Bioinformatics.,. 217. (2007). https://digitalcommons.wustl.edu/open_access_pubs/208 This Open Access Publication is brought to you for free and open access by Digital Commons@Becker. It has been accepted for inclusion in Open Access Publications by an authorized administrator of Digital Commons@Becker. For more information, please contact [email protected]. Authors Nathan Salomonis, Kristina Hanspers, Alexander C. Zambon, Karen Vranizan, Steven C. Lawlor, Kam D. Dahlquist, Scott .W Doniger, Josh Stuart, Bruce R. Conklin, and Alexander R. Pico This open access publication is available at Digital Commons@Becker: https://digitalcommons.wustl.edu/open_access_pubs/208 -

Genmapp 2: New Features and Resources for Pathway Analysis Nathan Salomonis Gladstone Institute of Cardiovascular Disease
Washington University School of Medicine Digital Commons@Becker Open Access Publications 2007 GenMAPP 2: New features and resources for pathway analysis Nathan Salomonis Gladstone Institute of Cardiovascular Disease Kristina Hanspers Gladstone Institute of Cardiovascular Disease Alexander C. Zambon Gladstone Institute of Cardiovascular Disease Karen Vranizan Gladstone Institute of Cardiovascular Disease Steven C. Lawlor Gladstone Institute of Cardiovascular Disease See next page for additional authors Follow this and additional works at: http://digitalcommons.wustl.edu/open_access_pubs Part of the Medicine and Health Sciences Commons Recommended Citation Salomonis, Nathan; Hanspers, Kristina; Zambon, Alexander C.; Vranizan, Karen; Lawlor, Steven C.; Dahlquist, Kam D.; Doniger, Scott .;W Stuart, Josh; Conklin, Bruce R.; and Pico, Alexander R., ,"GenMAPP 2: New features and resources for pathway analysis." BMC Bioinformatics.8,. 217. (2007). http://digitalcommons.wustl.edu/open_access_pubs/208 This Open Access Publication is brought to you for free and open access by Digital Commons@Becker. It has been accepted for inclusion in Open Access Publications by an authorized administrator of Digital Commons@Becker. For more information, please contact [email protected]. Authors Nathan Salomonis, Kristina Hanspers, Alexander C. Zambon, Karen Vranizan, Steven C. Lawlor, Kam D. Dahlquist, Scott .W Doniger, Josh Stuart, Bruce R. Conklin, and Alexander R. Pico This open access publication is available at Digital Commons@Becker: http://digitalcommons.wustl.edu/open_access_pubs/208 -

Transcriptomic Uniqueness and Commonality of the Ion Channels and Transporters in the Four Heart Chambers Sanda Iacobas1, Bogdan Amuzescu2 & Dumitru A
www.nature.com/scientificreports OPEN Transcriptomic uniqueness and commonality of the ion channels and transporters in the four heart chambers Sanda Iacobas1, Bogdan Amuzescu2 & Dumitru A. Iacobas3,4* Myocardium transcriptomes of left and right atria and ventricles from four adult male C57Bl/6j mice were profled with Agilent microarrays to identify the diferences responsible for the distinct functional roles of the four heart chambers. Female mice were not investigated owing to their transcriptome dependence on the estrous cycle phase. Out of the quantifed 16,886 unigenes, 15.76% on the left side and 16.5% on the right side exhibited diferential expression between the atrium and the ventricle, while 5.8% of genes were diferently expressed between the two atria and only 1.2% between the two ventricles. The study revealed also chamber diferences in gene expression control and coordination. We analyzed ion channels and transporters, and genes within the cardiac muscle contraction, oxidative phosphorylation, glycolysis/gluconeogenesis, calcium and adrenergic signaling pathways. Interestingly, while expression of Ank2 oscillates in phase with all 27 quantifed binding partners in the left ventricle, the percentage of in-phase oscillating partners of Ank2 is 15% and 37% in the left and right atria and 74% in the right ventricle. The analysis indicated high interventricular synchrony of the ion channels expressions and the substantially lower synchrony between the two atria and between the atrium and the ventricle from the same side. Starting with crocodilians, the heart pumps the blood through the pulmonary circulation and the systemic cir- culation by the coordinated rhythmic contractions of its upper lef and right atria (LA, RA) and lower lef and right ventricles (LV, RV). -

1471-2105-8-217.Pdf
BMC Bioinformatics BioMed Central Software Open Access GenMAPP 2: new features and resources for pathway analysis Nathan Salomonis1,2, Kristina Hanspers1, Alexander C Zambon1, Karen Vranizan1,3, Steven C Lawlor1, Kam D Dahlquist4, Scott W Doniger5, Josh Stuart6, Bruce R Conklin1,2,7,8 and Alexander R Pico*1 Address: 1Gladstone Institute of Cardiovascular Disease, 1650 Owens Street, San Francisco, CA 94158 USA, 2Pharmaceutical Sciences and Pharmacogenomics Graduate Program, University of California, 513 Parnassus Avenue, San Francisco, CA 94143, USA, 3Functional Genomics Laboratory, University of California, Berkeley, CA 94720 USA, 4Department of Biology, Loyola Marymount University, 1 LMU Drive, MS 8220, Los Angeles, CA 90045 USA, 5Computational Biology Graduate Program, Washington University School of Medicine, St. Louis, MO 63108 USA, 6Department of Biomolecular Engineering, University of California, Santa Cruz, CA 95064 USA, 7Department of Medicine, University of California, San Francisco, CA 94143 USA and 8Department of Molecular and Cellular Pharmacology, University of California, San Francisco, CA 94143 USA Email: Nathan Salomonis - [email protected]; Kristina Hanspers - [email protected]; Alexander C Zambon - [email protected]; Karen Vranizan - [email protected]; Steven C Lawlor - [email protected]; Kam D Dahlquist - [email protected]; Scott W Doniger - [email protected]; Josh Stuart - [email protected]; Bruce R Conklin - [email protected]; Alexander R Pico* - [email protected] * Corresponding author Published: 24 June 2007 Received: 16 November 2006 Accepted: 24 June 2007 BMC Bioinformatics 2007, 8:217 doi:10.1186/1471-2105-8-217 This article is available from: http://www.biomedcentral.com/1471-2105/8/217 © 2007 Salomonis et al; licensee BioMed Central Ltd. -

Chromatin Remodelling Complex Dosage Modulates Transcription Factor Function in Heart Development
ARTICLE Received 5 Aug 2010 | Accepted 11 Jan 2011 | Published 8 Feb 2011 DOI: 10.1038/ncomms1187 Chromatin remodelling complex dosage modulates transcription factor function in heart development Jun K. Takeuchi1,2,*, Xin Lou3,*, Jeffrey M. Alexander1,4, Hiroe Sugizaki2, Paul Delgado-Olguín1, Alisha K. Holloway1, Alessandro D. Mori1, John N. Wylie1, Chantilly Munson5,6, Yonghong Zhu3, Yu-Qing Zhou7, Ru-Fang Yeh8, R. Mark Henkelman7,9, Richard P. Harvey10,11, Daniel Metzger12, Pierre Chambon12, Didier Y. R. Stainier4,5,6, Katherine S. Pollard1,8, Ian C. Scott3,13 & Benoit G. Bruneau1,4,5,14 Dominant mutations in cardiac transcription factor genes cause human inherited congenital heart defects (CHDs); however, their molecular basis is not understood. Interactions between transcription factors and the Brg1/Brm-associated factor (BAF) chromatin remodelling complex suggest potential mechanisms; however, the role of BAF complexes in cardiogenesis is not known. In this study, we show that dosage of Brg1 is critical for mouse and zebrafish cardiogenesis. Disrupting the balance between Brg1 and disease-causing cardiac transcription factors, including Tbx5, Tbx20 and Nkx2–5, causes severe cardiac anomalies, revealing an essential allelic balance between Brg1 and these cardiac transcription factor genes. This suggests that the relative levels of transcription factors and BAF complexes are important for heart development, which is supported by reduced occupancy of Brg1 at cardiac gene promoters in Tbx5 haploinsufficient hearts. Our results reveal complex dosage-sensitive interdependence between transcription factors and BAF complexes, providing a potential mechanism underlying transcription factor haploinsufficiency, with implications for multigenic inheritance of CHDs. 1 Gladstone Institute of Cardiovascular Disease, San Francisco, California 94158, USA. -

Endocrine System Local Gene Expression
Copyright 2008 By Nathan G. Salomonis ii Acknowledgments Publication Reprints The text in chapter 2 of this dissertation contains a reprint of materials as it appears in: Salomonis N, Hanspers K, Zambon AC, Vranizan K, Lawlor SC, Dahlquist KD, Doniger SW, Stuart J, Conklin BR, Pico AR. GenMAPP 2: new features and resources for pathway analysis. BMC Bioinformatics. 2007 Jun 24;8:218. The co-authors listed in this publication co-wrote the manuscript (AP and KH) and provided critical feedback (see detailed contributions at the end of chapter 2). The text in chapter 3 of this dissertation contains a reprint of materials as it appears in: Salomonis N, Cotte N, Zambon AC, Pollard KS, Vranizan K, Doniger SW, Dolganov G, Conklin BR. Identifying genetic networks underlying myometrial transition to labor. Genome Biol. 2005;6(2):R12. Epub 2005 Jan 28. The co-authors listed in this publication developed the hierarchical clustering method (KP), co-designed the study (NC, AZ, BC), provided statistical guidance (KV), co- contributed to GenMAPP 2.0 (SD) and performed quantitative mRNA analyses (GD). The text of this dissertation contains a reproduction of a figure from: Yeo G, Holste D, Kreiman G, Burge CB. Variation in alternative splicing across human tissues. Genome Biol. 2004;5(10):R74. Epub 2004 Sep 13. The reproduction was taken without permission (chapter 1), figure 1.3. iii Personal Acknowledgments The achievements of this doctoral degree are to a large degree possible due to the contribution, feedback and support of many individuals. To all of you that helped, I am extremely grateful for your support. -

Systematic and Integrative Analysis of Proteomic Data Using Bioinformatics Tools
(IJACSA) International Journal of Advanced Computer Science and Applications, Vol. 2, No. 5, 2011 Systematic and Integrative Analysis of Proteomic Data using Bioinformatics Tools Rashmi Rameshwari Dr. T. V. Prasad Asst. Professor, Dept. of Biotechnology, Dean (R&D), Lingaya‟s University, Manav Rachna International University, Faridabad, India Faridabad, India Abstract— The analysis and interpretation of relationships or prediction. Expression and interaction experiments tend to between biological molecules is done with the help of networks. be on such a large scale that it is difficult to analyze them, or Networks are used ubiquitously throughout biology to represent indeed grasp the meaning of the results of any analysis. Visual the relationships between genes and gene products. Network representation of such large and scattered quantities of data models have facilitated a shift from the study of evolutionary allows trends that are difficult to pinpoint numerically to stand conservation between individual gene and gene products towards out and provide insight into specific avenues of molecular the study of conservation at the level of pathways and complexes. functions and interactions that may be worth exploring first out Recent work has revealed much about chemical reactions inside of the bunch, either through confirmation or rejection and then hundreds of organisms as well as universal characteristics of later of significance or insignificance to the research problem at metabolic networks, which shed light on the evolution of the hand. With a few recent exceptions, visualization tools were networks. However, characteristics of individual metabolites have been neglected in this network. The current paper provides not designed with the intent of being used for analysis so much an overview of bioinformatics software used in visualization of as to show the workings of a molecular system more clearly. -

Mappfinder: Using Gene Ontology and Genmapp to Create a Global
Method Open Access MAPPFinder: using Gene Ontology and GenMAPP to create a comment global gene-expression profile from microarray data Scott W Doniger*, Nathan Salomonis*, Kam D Dahlquist*†, Karen Vranizan*‡, Steven C Lawlor* and Bruce R Conklin*†§ Addresses: *Gladstone Institute of Cardiovascular Disease, University of California, San Francisco, CA 94141-9100, USA. †Cardiovascular Research Institute, and §Departments of Medicine and Cellular and Molecular Pharmacology, University of California, San Francisco, ‡ CA 94143, USA. Functional Genomics Lab, University of California, Berkeley, CA 94720, USA. reviews Correspondence: Bruce R Conklin. E-mail: [email protected] Published: 6 January 2003 Received: 11 September 2002 Revised: 8 October 2002 Genome Biology 2003, 4:R7 Accepted: 8 November 2002 The electronic version of this article is the complete one and can be found online at http://genomebiology.com/2003/4/1/R7 reports © 2003 Doniger et al.; licensee BioMed Central Ltd. This is an Open Access article: verbatim copying and redistribution of this article are permitted in all media for any purpose, provided this notice is preserved along with the article's original URL. Abstract deposited research MAPPFinder is a tool that creates a global gene-expression profile across all areas of biology by integrating the annotations of the Gene Ontology (GO) Project with the free software package GenMAPP (http://www.GenMAPP.org). The results are displayed in a searchable browser, allowing the user to rapidly identify GO terms with over-represented numbers of gene- expression changes. Clicking on GO terms generates GenMAPP graphical files where gene relationships can be explored, annotated, and files can be freely exchanged. -

Microarray Data Analysis Tool (Mat)
MICROARRAY DATA ANALYSIS TOOL (MAT) A Thesis Presented to The Graduate Faculty of The University of Akron In Partial Fulfillment of the Requirements for the Degree Master of Science Sudarshan Selvaraja December, 2008 MICROARRAY DATA ANALYSIS TOOL (MAT) Sudarshan Selvaraja Thesis Approved: Accepted: _______________________ _______________________ Advisor Department Chair Dr. Zhong-Hui Duan Dr. Wolfgang Pelz _______________________ _______________________ Committee Member Dean of the College Dr. Yingcai Xiao Dr. Ronald F. Levant _______________________ _______________________ Committee Member Dean of the Graduate School Dr. Xuan-Hien Dang Dr. George R. Newkome _______________________ Date ii ABSTRACT Microarray is a technology that has been widely used by the biologists to probe the presence of genes in a sample of DNA or RNA. Using the technology, the oligonucleotide probes can be massively parallel immobilized on a microarray chip. It allows the biologists to check the expression levels of thousands of genes together. This thesis develops a software system that includes a database repository to store different microarray datasets and a microarray data analysis tool for analyzing the stored data. The repository currently allows datasets of GenepixPro format to be deposited, although it can be expanded to include datasets of other formats. The user interface of the repository allows users conveniently upload data files and perform preferred data preprocessing and analysis. The analysis methods implemented includes the traditional k-nearest neighbor (kNN) methods and two new kNN methods developed in this study. Additional analysis methods can be added by future developers. The system was tested using a set of microRNA gene expression data. The design and implementation of the software tool are presented in the thesis along with the testing results from the microRNA dataset. -

A Computational Genomics Perspective
Abstract of thesis entitled Understanding the Pathogenic Fungus Penicillium marne®ei:A Computational Genomics Perspective by James J. Cai for the degree of Doctor of Philosophy at The University of Hong Kong in May 2006 Penicillium marne®ei, a thermally dimorphic fungus that alternates be- tween a ¯lamentous and a yeast growth form in response to changes in its environmental temperature, has become an emerging fungal pathogen endemic in Southeast Asia. De¯ning the genomics of P. marne®ei will provide a better understanding of the fungus. This thesis reports the draft sequence of the P. marne®ei genome as- sembled from 6.6 coverage of the genome through whole genome shotgun sequencing. The 31 Mb genome obtained from the assembly contains 10,060 protein-coding genes. The complete mitochondrial genome is 35 kb long and its gene content and gene order are very similar to that of Aspergillus. An annotation system and P. marne®ei genome database (PMGD) were developed to allow a preliminary annotation of the se- quences and provide an intuitive graphic interface to give curators and users ready access to the annotation and the underlying evidence, and a Matlab-based software package, MBEToolbox, was developed for data analysis in phylogenetics and comparative genomics. A well-designed and structured annotation system and powerful sequence analysis software are essential requirements for the success of large-scale genome analysis projects. Analysis of the gene set of P. marne®ei provided insights into the adaptations required by a fungus to cause disease. The genome encodes a diverse set of putative virulence genes such as proteinase, phospholi- pase, metacaspase and agglutinin, which may enable the fungus to adhere to, colonise and invade the host, adapt to the tissue environment, and avoid the host's humoral and cellular defences of the innate and adaptive immune responses.